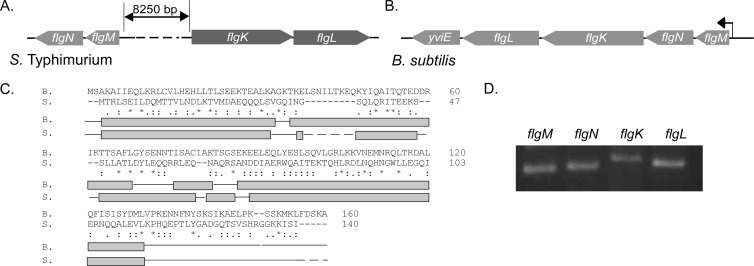
FIG 1.
Primary-sequence and secondary-structure comparison of B. subtilis FlgN with S. Typhimurium FlgN. Schematic representations of the chromosomal regions surrounding S. Typhimurium flgN (A) and B. subtilis flgN (B) are shown. Arrows represent open reading frames, with the direction of the arrow indicating the direction of the open reading frame. The bent arrow represents the promoter located before the flgM coding region. (C) Primary-sequence alignment and secondary-structure comparison of Bs-flgN and ST-flgN. The primary amino acid sequence of FlgN from B. subtilis 168 (B) was aligned with that of flgN from S. Typhimurium (S). For primary-sequence alignments, an asterisk indicates a fully conserved amino acid, a colon indicates a highly conserved amino acid, and a period indicates a weakly conserved amino acid. Gaps indicate no homology. Dashed lines indicate a break in the sequence. For secondary-structure alignments light gray boxes indicate α-helices, solid lines indicate coiled coils, and dashed lines indicate a break in the sequence alignment. (D) RT-PCR analysis of cotranscription of flgM, flgN, flgL, and flgK. Regions of DNA internal to flgM, flgN, flgK, and flgL were amplified from cDNA generated using a primer specific to the gene 3′ proximal to flgL, using RNA from the wild-type strain (NCIB3610).