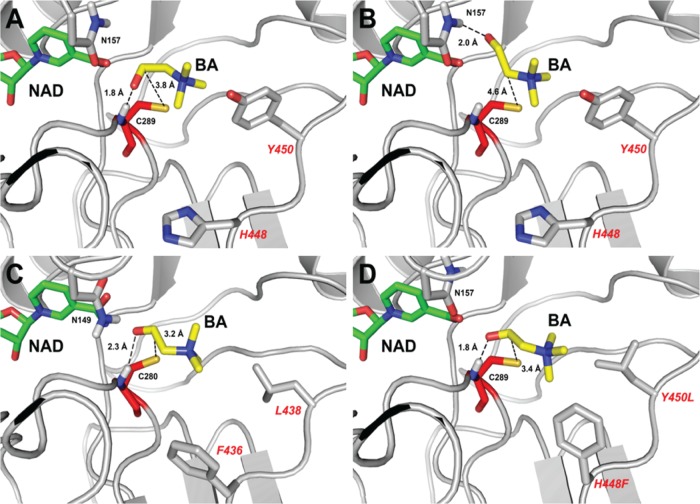
FIG 7.
Structural basis of BetB inhibition by BA: productive and nonproductive binding of BA. Docking simulation of BA binding in the active sites of wild-type BetB (A, B), YdcW (C), and the homology model of the double mutant protein H448F/Y450L (D). Panels A, C, and D represent the productive binding mode of BA, with a hydrogen bond formed between the carbonyl group of BA and the backbone nitrogen atom of Cys289 (in BetB). In contrast, panel B presents BA in a nonproductive binding mode with similar binding affinity and a hydrogen bond formed between the carbonyl group of BA and the amide group of Asn157. The amino acid side chains and ligands are shown as sticks and labeled. The residues of the double mutant protein H448F/Y450L are labeled in a larger font.