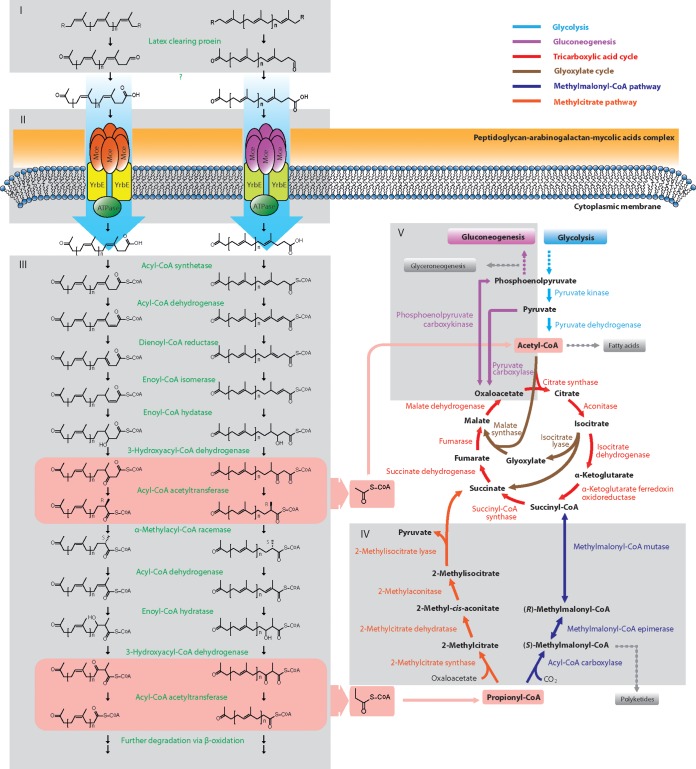
FIG 1.
Proposed metabolic pathway of polyisoprene degradation in N. nova SH22a. The six gray blocks labeled by roman numbers represent the main parts of the proposed degradation pathways. I, initial oxidative cleavage of polyisoprene chains (indirect evidence from other studies [13, 14, 51]); II, import of oligoisoprenes (direct evidence from transposon-induced mutants of Mce in strain SH22a [this work] and also in the closely related strain G. polyisoprenivorans VH2 [14]); III, β-oxidation (direct evidence from transposon-induced mutants, deletion mutants, and complementation experiments for α-methylacyl-CoA racemase in strain SH22a [15], and also indirect evidence from other studies [62, 98]); IV, propionyl-CoA metabolism (direct evidence from transposon-induced mutants in strain SH22a [this work]); V, gluconeogenesis (direct evidence from transposon-induced mutants, deletion mutants, and complementation experiments for phosphoenolpyruvate carboxykinase [this work]). The two pink bars show the steps alternately releasing degradation products (acetyl-CoA and propionyl-CoA) of β-oxidation.