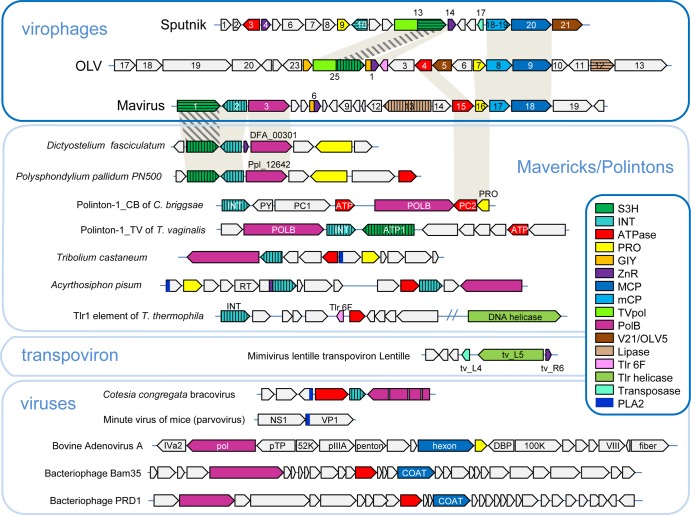
FIG 11.
Comparison of the genome architectures of virophages and polinton-like transposable elements. Homologous genes are color coded. Different hatching patterns are used to mark nonorthologous primase-helicase, integrase, and lipase genes. Homologous regions are shaded. Reference sequences were extracted from GenBank, using the following accession numbers: Dictyostelium fasciculatum, GI:328871053; Polysphondylium pallidum, GI:281202948; Tribolium castaneum, GI:58197573; Acyrthosiphon pisum, GI:156713484; Mimivirus lentille transpoviron Lentille, GI:374110342; Cotesia congregata bracovirus, GI:326937614; minute virus of mice, GI:9626993; bovine adenovirus A, GI:52801677; bacteriophage Bam35, GI:38640293; and bacteriophage PRD1, GI:159192286. Some sequences were extracted from Repbase (Polinton-1_CB and Polinton-1_TV [156, 202]). PLA2, phospholipase A2 domain of the parvovirus capsid protein. Other color key abbreviations are the same as those used throughout the text. (Adapted from reference 32, published under a Creative Commons license.)