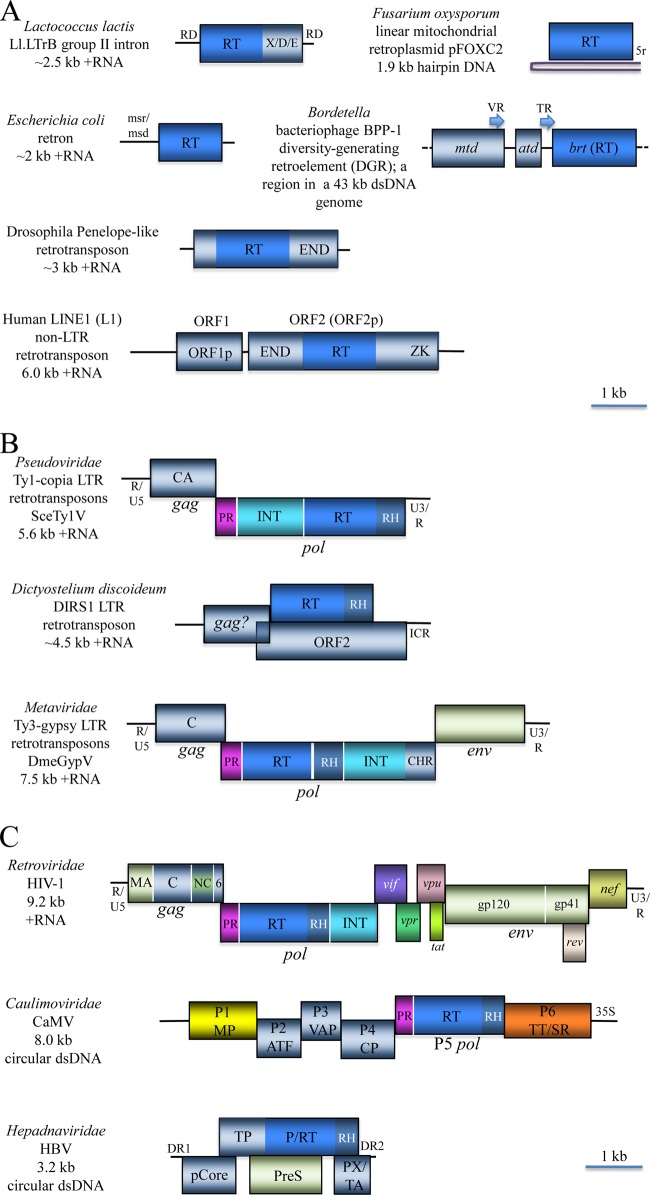
FIG 6.
Representative genome architectures of retroelements and the derivative retroviruses. (A) Prokaryotic and eukaryotic capsidless retroelements. Group II introns are scattered in genomes of diverse bacteria and some archaea, as well as mitochondrial and chloroplast genomes of many eukaryotes. Retrons are typical of bacteria, whereas Penelope-like and non-LTR retrotransposons are widespread in diverse eukaryotes. The diversity-generating retroelements (DGR) are present in a narrow range of tailed DNA bacteriophages and in some bacteria. Linear mitochondrial retroplasmids are present in some fungi. RD, RNA domains involved in the splicing of intron RNA; X/D/E, maturase, DNA binding, and endonuclease domains, respectively, of the intron-encoded protein; msr/msd, regions encoding RNA and DNA components, respectively, of the satellite msDNA; 5r, a telomere-like iteration of a 5-nucleotide sequence; VR, variable repeat; TR, template repeat; mtd, major tropism determinant; atd, accessory tropism determinant; brt, bacteriophage reverse transcriptase; LINE, long interspersed nucleotide elements; ORF1p and ORF2p, ORF1 and 2 proteins; END, endonuclease; ZK, zinc knuckle. (B) The LTR (long terminal repeat) retrotransposons are ubiquitous in eukaryotes. Because many of them form primarily noninfectious, virion-like particles encoded by the gag (group-specific antigen) and env (envelope) ORFs, two classes of these retrotransposons are recognized as viral families Metaviridae and Pseudoviridae. The pol (polymerase) ORF encodes a complete or partial complement of the aspartate protease (PR), reverse transcriptase (RT), RNase H (RH), and integrase (INT) domains and, in Metaviridae, a chromodomain (CHR). The sites of Pol processing by PR are shown as vertical white lines. ICR, internal complementarity region. Viral name acronyms: DmeGypV, Drosophila melanogaster gypsy virus; SceTy1V, Saccharomyces cerevisiae Ty1 virus. (C) Reverse-transcribing (retroid) viruses. The genomes are shown as RNA or primarily double-stranded DNA that is circular but rendered linear for the sake of comparison. In HIV-1, both gag and pol are processed (vertical white lines) by PR, whereas env is processed by the host proteases. MA, matrix protein; C, capsid protein; NC, nucleocapsid; 6, 6-kDa protein; vif, vpr, vpu, tat, rev, and nef, regulatory proteins encoded by spliced mRNAs (only the main parts of the coding regions are shown); gp120 and gp41, the 120- (surface) and 41-kDa (transmembrane) glycoproteins; ATF, aphid transmission factor; VAP, virion-associated protein; CP, capsid protein; TT/SR, translation trans-activator/suppressor of RNA interference; 35S, 35S RNA polymerase Pol II promoter; pCore, capsid (core) protein; TP, terminal protein; P, polymerase; PreS, pre-surface protein (envelope); PX/TA, protein X/transcription activator; DR1 and DR2, direct repeat sequences; HIV-1, Human immunodeficiency virus 1; CaMV, Cauliflower mosaic virus; HBV, Hepatitis B virus.