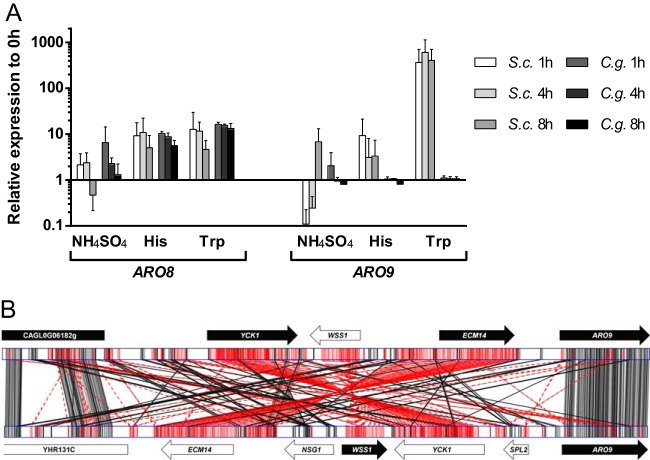
FIG 5.
(A) Relative expression of ARO8 and ARO9 after transfer from complex medium to minimal medium containing ammonium sulfate, histidine, or tryptophan as the sole source of nitrogen. In both S. cerevisiae (S.c.) and C. glabrata (C.g.), ARO8 expression was upregulated compared to its expression in the preculture at all time points when the amino acids were offered. There was no regulation observable for ARO9 in C. glabrata, while S. cerevisiae very strongly upregulated ARO9 when tryptophan was present. The error bars indicate standard deviations. (B) Genomic-locus alignment of the ARO9 gene and 10-kbp upstream regions in C. glabrata (top) and S. cerevisiae (bottom). An inversion event was detected encompassing the upstream genes YCK1, WSS1, and ECM14. The genes CAGL0G06182 and YHR131C are homologues with identical orientations with respect to the ARO9 genes. The solid arrows indicate ORFs with the same orientation as ARO9, and the open arrows indicate opposite orientations. Alignment was performed with GATA (22). Black lines indicate alignments in the same orientation (+/+), and red lines indicate those in inverse orientation (+/−). Darker shadings indicate better alignment scores.