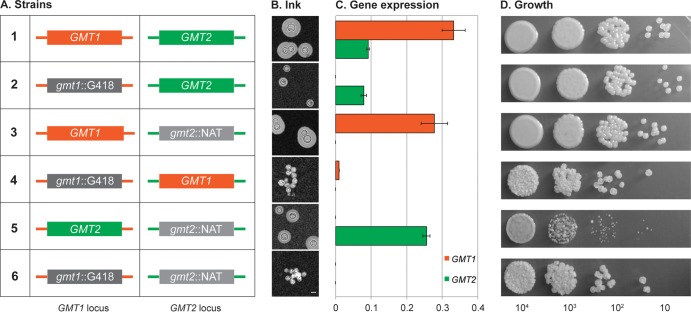
FIG 8.
Genomic location does not explain the functional differences between GMT1 and GMT2. (A) Schematic representation of the GMT strain variants listed at left, indicating the coding sequence present at each locus (colored boxes): orange, GMT1; green, GMT2; dark gray, Geneticin resistance marker (G418) replacing GMT1 (gmt1::G418); light gray, nourseothricin resistance marker (NAT) replacing GMT2 (gmt2::NAT). Promoter and terminator sequences of each locus (colored lines) were not altered. Strains: 1, wild type; 2, gmt1 mutant; 3, gmt2 mutant; 4 and 5, new “swapped” strains; 6, gmt1 gmt2 double mutant. (B) The capsule thickness of each strain under capsule-inducing conditions was examined with India ink as described in Materials and Methods. Scale bar, 5 μm. (C) GMT expression levels under capsule-inducing conditions determined by RT-PCR, plotted relative to ACT1. (D) Growth on rich medium. Aliquots (5 μl) of a stock cell suspension (2 × 106 cells/ml) and three 10-fold serial dilutions were spotted on YPD plates and grown for 1 day at 30°C.