FIG 2.
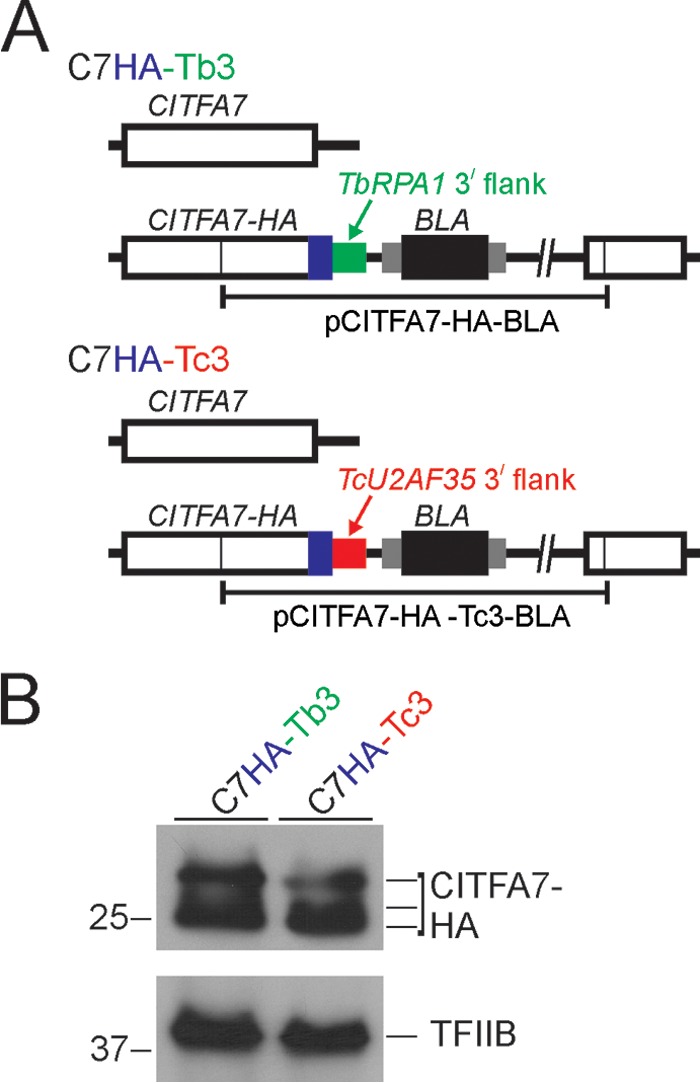
The 3′ gene flank and UTR of T. cruzi TcU2AF35 are functional in T. brucei. (A) Schematic outline (not to scale) of the CITFA7 locus in the control cell line C7HA-Tb3 and in the cell line C7HA-Tc3. In both cell lines, the HA tag sequence was fused to the CITFA7 coding region by targeted integration of the specified linearized plasmid into one CITFA7 allele. The difference is that in C7HA-Tb3 and C7HA-Tc3 cells, the manipulated CITFA7 allele is under the control of the T. brucei RPA1 (green) and the heterologous TcU2AF35 (red) 3′ gene flanks, respectively. The CITFA7 coding region, the HA tag, and the BLA sequences are indicated by open, blue, and black boxes, respectively. The smaller gray boxes surrounding BLA represent T. brucei gene flanks providing RNA-processing signals. (B) Immunoblot of C7HA-Tb3 and C7HA-Tc3 whole-cell lysates detecting CITFA7-HA with a monoclonal anti-HA antibody and, as a loading control, the transcription factor TFIIB with an anti-TFIIB polyclonal immune serum. Note that, due to phosphorylation, CITFA7 separates into multiple bands.
