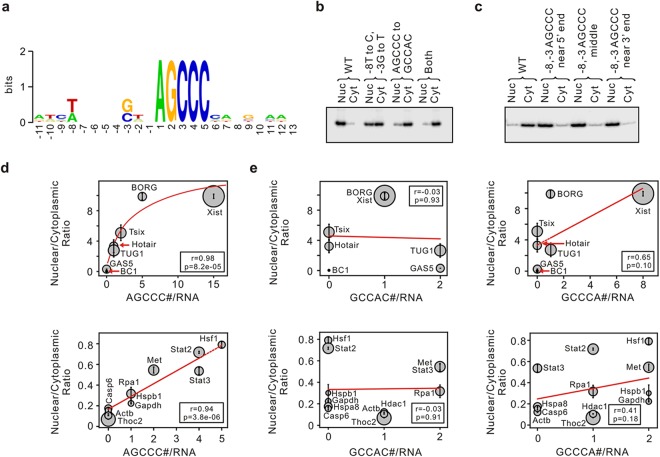
FIG 6.
A short RNA motif mediates the localization of RNAs to the nucleus. (a) Information content of the nucleotide positions within and around the AGCCC motif. (b) Mutation of the AGCCC sequence or the nucleotides at positions −3 and −8 leads to loss of nuclear localization of Frag2 RNA. The identity of the mutation tested is shown above each set of samples. Nuc and Cyt refer to nuclear and cytoplasmic fractions, respectively. WT, unmodified Frag2 transcript. (c) Insertion of a single copy of the motif near the 5′ or 3′ end or the middle of the Frag5-Reverse+complement RNA results in nuclear localization. WT, unmodified Frag5-Reverse+complement RNA. (d) The copy number of −8 (A/T), −3 (G/C), AGCCC motif per transcript shows a direct correlation with nuclear localization in both lncRNAs (top) and mRNAs (bottom). The correlation coefficient (r) and two-tailed P values (p) for the log (top) and linear (bottom) fits are shown. The size of the circle marking each transcript is proportional to the length of the RNA. The center of each circle corresponds to the average value of three independent experiments, with vertical lines (error bars) corresponding two standard deviations. The identity of each RNA is shown. (e) Analysis similar to that in panel d with two scrambled pentamers replacing AGCCC indicating the specificity of the correlation observed with AGCCC.