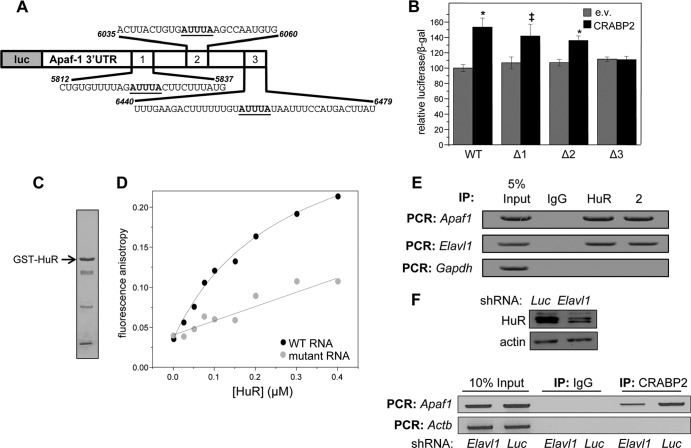
FIG 3.
HuR mediates the association of CRABP2 with mRNA. (A) Diagram of the luciferase reporter harboring the Apaf1 3′ UTR. Putative ARE are underlined. (B) M2−/− cells were transfected with an e.v. or a vector encoding CRABP2 and the luciferase reporter harboring the Apaf1 3′ UTR or counterparts lacking the indicated putative HuR binding sites (Δ1, Δ2, and Δ3). β-Galactosidase was used as a transfection control. Data were normalized to luciferase activity in cells transfected with e.v. and the WT luciferase reporter. Data are means ± standard errors of the means (n = 3). *, P ≤ 0.01, and ‡, P = 0.025, both versus e.v. control by a two-tailed Student t test. (C) Coomassie blue-stained gels visualizing recombinant, bacterially expressed, and purified GST-HuR. (D) Fluorescein-labeled RNA containing 39 bases corresponding to site 3 in the Apaf-1 3′ UTR (WT RNA) or fluorescein-labeled RNA with the AUUUA of site 3 deleted (mutant RNA) was titrated with recombinant GST-tagged HuR. Progress of titrations was followed by monitoring the increase in the fluorescence anisotropy of the labeled RNA (λexcitation = 494 nm; λemission = 518 nm). Data representative of 3 independent experiments are shown. (E) HuR and CRABP2 were immunoprecipitated from lysates of M2−/− cells stably expressing EGFP-CRABP2 (Fig. 1A). Apaf1 and Elavl1 mRNAs that coprecipitated with the proteins were assessed by semiquantitative PCR. (F) M2−/− cells that stably overexpress CRABP2 were infected with lentiviruses harboring vectors encoding shRNA targeting Elavl1 (shElavl1) or luciferase (shLuc), and stable cell lines were generated. (Top) Immunoblot demonstrating HuR levels in cells expressing shLuc or shElavl1. Actin was used as loading control. (Bottom) CRABP2 was immunoprecipitated and RNAs that coprecipitated with the protein were assessed by semiquantitative PCR.