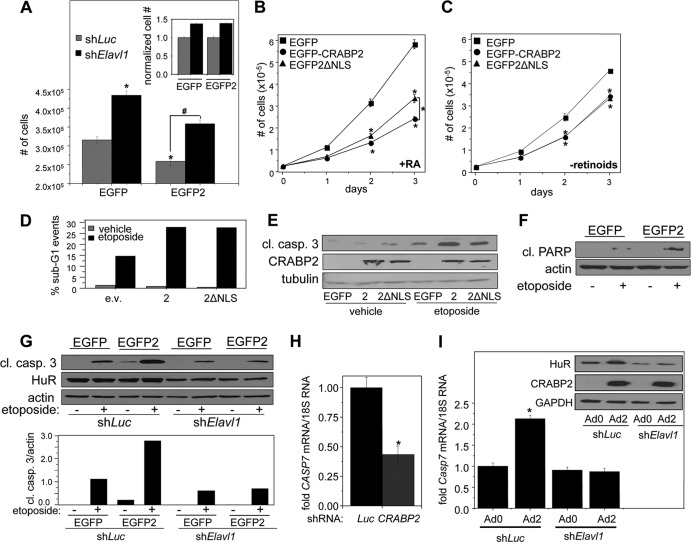
FIG 6.
CRABP2 enhances apoptosis by cooperating with HuR. In all experiments, cells were cultured in delipidated medium for 48 h to deplete retinoid stores. (A) M2−/− cells stably expressing EGFP or EGFP-CRABP2 and harboring shRNAs targeting Elavl1 (shElavl1) or luciferase (shLuc) were plated at 35,000 cell/well and counted after 4 days of growth. Data are means ± standard errors of the means (n = 3). Statistical analysis was carried out by one-way analysis of variance and Bonferroni's post hoc test. *, P < 0.01 versus cells expressing EGFP and shLuc; #, P < 0.01 versus cells expressing EGFP and shElavl1. (Inset) The number of shElavl1-expressing cells was normalized to the corresponding shLuc control. (B) Growth of M2−/− cells stably overexpressing the indicated proteins in delipidated medium supplemented with 200 nM RA. Data are means ± standard errors of the means (n = 3). *, P < 0.01 versus EGFP control cells and, on day 3, P < 0.01 comparing cells expressing EGFP-CRABP2 versus EGFP-2ΔNLS. Analysis was carried out by one-way analysis of variance and Bonferroni's post hoc test. (C) Growth of M2−/− cells stably overexpressing the indicated proteins in delipidated medium. Data are means ± standard errors of the means (n = 3). *, P ≤ 0.01 versus control EGFP-expressing cells using one-way analysis of variance with Bonferroni's post hoc test. P > 0.1 for EGFP-CRABP2 versus EGFP-2ΔNLS on days 2 and 3, as calculated by two-tailed Student t test. (D and E) M2−/− cells were transfected with vectors encoding EGFP-CRABP2 or EGFP-CRABP2ΔNLS. Cells were treated with etoposide (10 μM for 48 h). (D) Apoptosis was evaluated by FACS to quantitate percentage of cells in G1. The experiment was repeated, with similar results. (E) Apoptosis was evaluated by using immunoblotting to monitor cleavage of caspase 3. The experiment was repeated, with similar results. (F) M2−/− cells were stably transfected with EGFP or EGFP-CRABP2 and treated with etoposide (10 μM for 48 h), and apoptosis was assessed by monitoring PARP cleavage by immunoblotting. (G) (Top) M2−/− cells stably expressing EGFP or EGFP-CRABP2 were infected with lentivirus harboring shRNA targeting either Elavl1 (shElavl1) or luciferase (shLuc). Cells were then treated with etoposide (10 μM for 48 h), and apoptosis was evaluated by immunoblotting monitoring caspase 3 cleavage. (Bottom) Quantitation of immunoblots. The experiment was repeated, with similar results. (H) MCF-7 cells were infected with lentiviruses containing vector harboring shRNA targeting CRABP2 (shCRABP2) or luciferase (shLuc). Three days postinfection, cells were harvested and the level of CASP7 mRNA was assessed by qPCR. *, P ≤ 0.01 versus shLuc, determined using two-tailed Student t test. See Fig. 1D and E for knockdown of CRABP2. (I) M2−/− cells were infected with lentiviruses containing vector harboring shRNA targeting Elavl1 (shElavl1) or luciferase (shLuc). Forty-eight hours later, cells were infected with control adenovirus (Ad0) or adenovirus encoding CRABP2 (Ad2). Forty-eight hours after adenoviral infection, RNA was extracted and Casp7 mRNA levels were assessed by qPCR. Data are means ± standard errors of the means (n = 3). *, P < 0.01 versus cells expressing shLuc and Ad0 by two-tailed Student t test. (Inset) Immunoblots demonstrating decreased expression of HuR in cells expressing shElavl1 and increased expression of CRABP2 upon infection with Ad2.