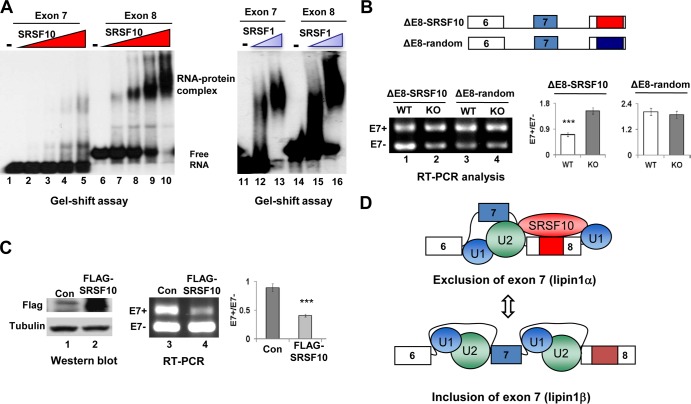
FIG 5.
Characteristics of SRSF10-regulated lipin1α/β splicing. (A) The indicated 32P-labeled RNAs were incubated with increasing amounts of recombinant His-SRSF10 (75 ng, 150 ng, 300 ng, and 600 ng), and complexes were resolved by nondenaturing PAGE (left). Each of the indicated RNA probes was incubated with 150 ng or 300 ng of recombinant SRSF1 and analyzed (right). (B) Three copies of SRSF10 consensus sequences or random control sequences were inserted downstream of the ΔE8 minigene to construct ΔE8-SRSF10 and ΔE8-random derivatives (top). F2/R2 primer pairs were used for in vivo splicing analysis, and quantification was performed as described for Fig. 4C (bottom). (C) Cotransfection of KO immortalized MEF cells with lipin1 minigene in the presence of either vector or FLAG-SRSF10 plasmid. Whole-cell lysates were prepared and analyzed by Western blotting using anti-FLAG and antitubulin antibodies (left). RNA was isolated and analyzed by RT-PCR, and quantification was performed as described for Fig. 4C (right). (D) Model for inhibition of lipin1 exon 7 inclusion by SRSF10. (Top) SRSF10 binding to lipin1 exon 8 containing the SRSF10 binding region (red box) stimulates or stabilizes interactions of general splicing factor U1 or U2 snRNPs at the nearby constitutive splice sites, thereby decreasing the competitiveness of the competing alternative sites and causing exon 7 skipping. (Bottom) In contrast, when binding of SRSF10 to exon 8 is disrupted, stabilization of U1 or U2 snRNPs at alternative splice sites for exon 7 is relatively stimulated, hence leading to exon 7 inclusion.