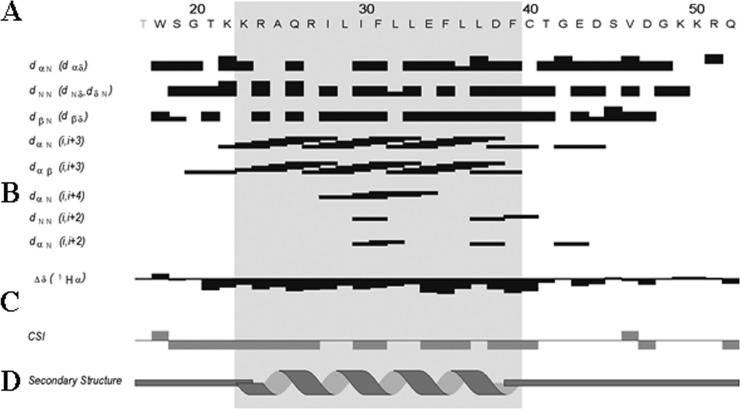
FIG 3.
Summary of sequential and short-range NOE connectivities for the agnoprotein peptide. (A) Amino acid sequence of the agnoprotein peptide. (B) Characteristic strong dNN (i, i + 1), dαN (i, i + 1), medium-range dαN (i, i + 2; i, i + 3; i, i + 4), and dαβ (i, i + 3) signals identified by TOCSY, DQF-COSY, and NOESY experiments. α, β, and N are the alpha, beta, and amide protons of each amino acid, respectively. dαβ, dNN, and dβN are the distances between alpha and beta, between amide and amide, and between beta and amide protons of two distinct residues, respectively. The thickness of the lines is related to the estimated intensity of the NOE (strong, medium, and weak). (C) The Δδ and CSI represented indicate the secondary structure formation according to the H-α chemical shift. (D) Schematic organization of the agnoprotein peptide, showing the secondary structures deduced from the NOESY spectra. Δδ is the chemical shift index (CSI) used to display and identify the location and the type of protein secondary structures using 1Hα chemical shift data, which are shifted upfield in helices and downfield in beta strands relative to their random coil values.