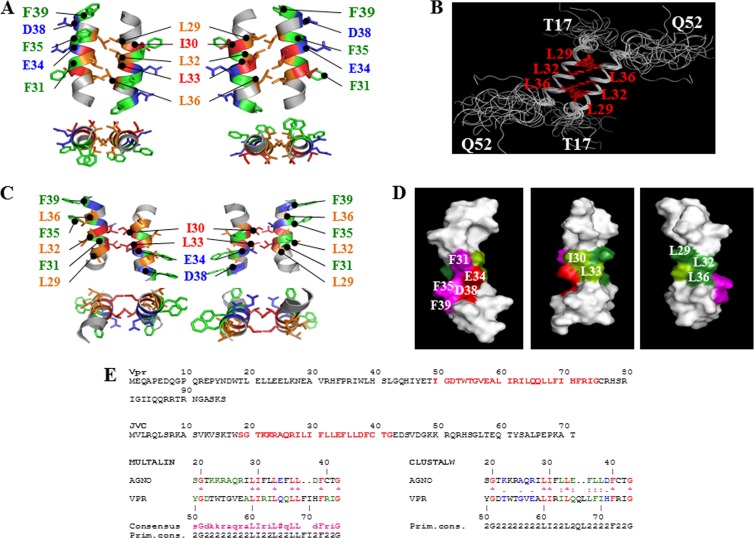
FIG 6.
Theoretical model of the dimeric structure of agnoprotein. (A) The energetically most favorable structure of agnoprotein calculated with X-PLOR (45, 46) is shown with the backbone in a ribbon representation and the side chains of the important hydrophobic and aromatic residues shown in stick representations. The structure is characterized by two antiparallel α-helical domains from residues K23 to F39 with an interface composed of residues, L29, L32, and L36 (orange). Residues I30 and L33 (dark orange) are perpendicular to this interface, and aromatic residues (green) are on the opposite face of the α-helix. The domain from residues K23 to F39 is well structured with good convergence, while the N- and C-terminal domains (not shown) are unstructured. (B) Seventeen different dimeric structures of the agnoprotein peptide were superimposed on the average structure. Hydrophobic amino acids (L29, L32, and L36) at the interface of the protein peptide are colored red. (C) An alternative theoretical model of the dimeric structure of agnoprotein. The energetically most favorable structure of agnoprotein was calculated using X-PLOR (45, 46), as described for panel A. (D) Surface-filling representation of the NMR structure of agnoprotein. The positions of the various amino acids in the α-helix are indicated. (E) Sequence alignment of Vpr and JCV agnoprotein within their dimerization domains. Primary sequences for both Vpr and JCV agnoprotein are shown. Sequences labeled in red for both proteins were aligned using the Multalin (77) and CLUSTALW (78) programs. The best correspondence in the alignment was found using Multalin when L29, I30, L33, L36, and L37 in the JCV agnoprotein corresponded, respectively, to L60, I61, L64, L67, and L68 in Vpr.