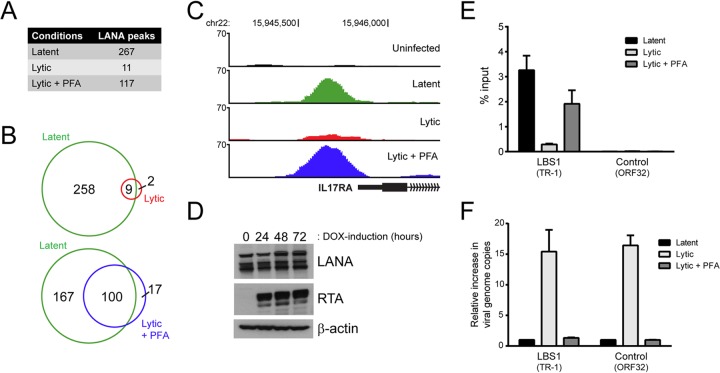
FIG 7.
The association of LANA with host DNA and viral TRs is impaired in lytically reactivated iSLK-219s. (A) Number of reproducible LANA peaks detected in latent, lytic, and PFA-treated lytic cells from two independent ChIP-seq experiments. (B) Venn diagrams illustrating the overlap between LANA-enriched regions detected in lytic and PFA-treated lytic cells compared to LANA-binding sites mapped in latent cells. (C) Screenshots from the UCSC browser (hg19) illustrating the loss of LANA enrichment within the promoter region of IL-17RA in lytically reactivated iSLK-219 cells. (D) The expression of LANA was examined by Western blotting in iSLK-219 cells that were mock induced or DOX induced for 24, 48, or 72 h. RTA and β-actin levels were assayed as controls of lytic reactivation and protein loading, respectively. (E) The enrichment of LANA at the viral LBS1 motif (TR-1) and at a control viral site (ORF32) in latent, lytic, and PFA-treated lytic cells was assessed by ChIP-qPCR. (F) The increase in viral genome copy number was determined by ChIP-qPCR using the input material from the ChIP experiment described in panel E. The amount of DNA fragment mapping to the indicated viral sites was compared to a control region on the host genome (GAPDH), and the results were normalized to those obtained in latent cells.