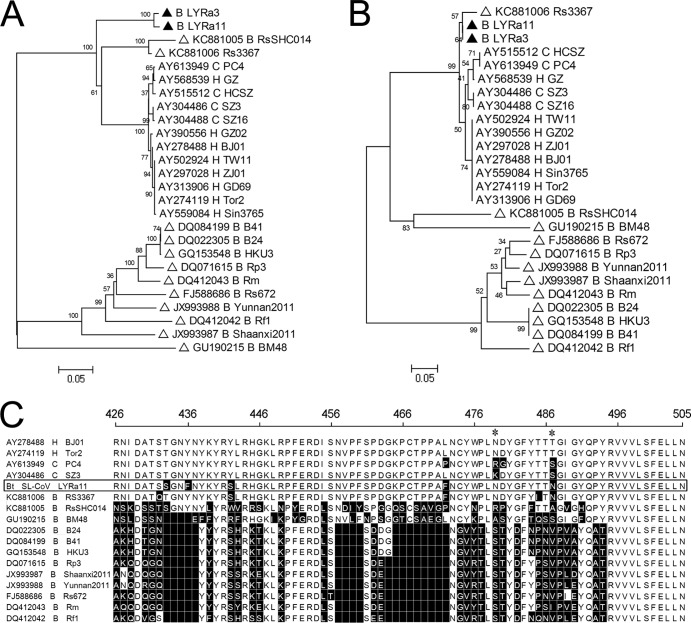
FIG 4.
Characterization of S1 domains of SARS and SARS-like CoVs. (A) Phylogenetic analysis of entire S1 amino acid sequences based on the maximum likelihood method; (B) phylogenetic analysis of RBD amino acid sequences based on the maximum likelihood method; (C) sequence comparison of entire RBMs of SARS CoVs, LYRa11 (boxed), and other closely related bat SARS-like CoVs. The sequences of SARS-like CoVs in this study are marked as filled triangles, with other bat SARS-like CoVs as open triangles. Middle letters: H, human SARS CoV; C, civet SARS CoV; B, bat SARS-like CoV. Amino acid (aa) positions refer to SARS CoV Tor2 (AY274119). Critical residues that play key roles in receptor binding are indicated with asterisks.