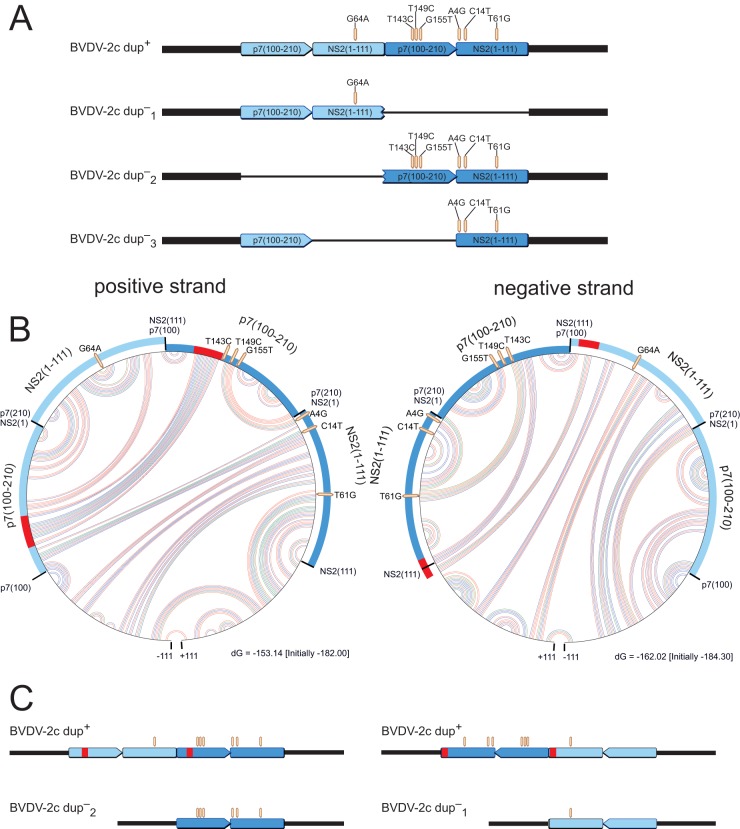
FIG 4.
Schema of theoretically possible dup− variants and potential RNA secondary structures facilitating their generation. (A) Schematic representation of theoretically possible dup− variants fitting the BVDV-2c prototype Potsdam 1600 that could be generated from dup+ genomes by deletions. Different shades of blue indicate different copies of the p7-NS2 region. (B) Circular graphs of RNA secondary structure predictions of the p7-NS2 region for the positive and negative strands. The outer circle represents the 5′-to-3′ sequence (clockwise). Arcs within the circle show base interactions (red, G-C; blue, A-U; and green, G-U). Along the circle, the modules of the duplicated region are highlighted. The sections marked in red represent interacting RNA stretches which bring corresponding regions of the two copies into close proximity, thereby providing an opportunity for the polymerase to circumvent exactly one copy. (C) Schematic straight representations of the circular parts of panel B, without base interactions.