FIG 6.
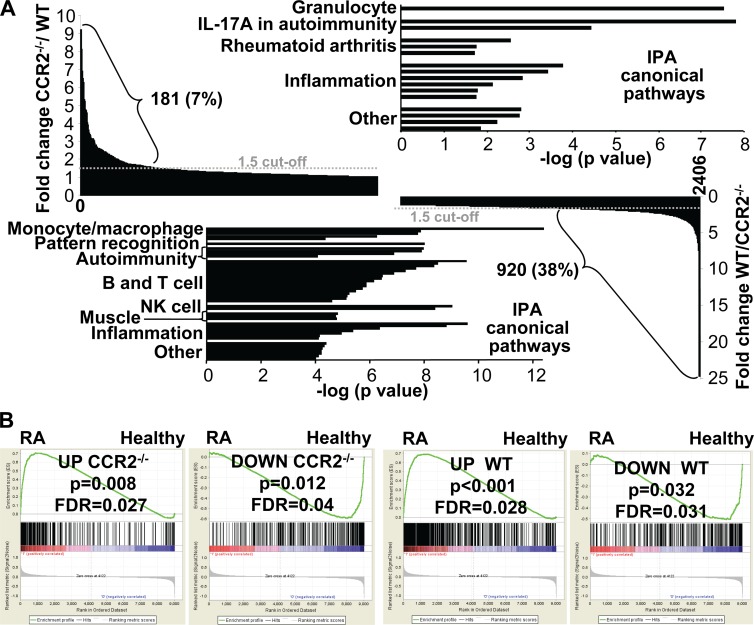
Microarray analysis of arthritic feet from WT and CCR2−/− mice. (A) Genes differentially expressed on day 7 postinfection (compared to day 0) in WT and CCR2−/− feet were determined by microarray analysis. A total of 2,406 differentially expressed genes were up- or downregulated in WT and/or CCR2−/− feet (primary data available upon request). For 181 of these genes, the ratio of the fold change in CCR2−/− mice to the fold change in WT mice was >1.5 (top left) (for 603 DEGs, this ratio was >1). For 920 of these genes, the ratio of the fold change in WT mice to the fold change in CCR2−/− mice was >1.5 (bottom right) (for 1,794 genes, this ratio was >1, and for 9 genes, this ratio was 1). IPAs of the 181 genes more induced in CCR2−/− feet (top right) and the 920 genes less induced in CCR2−/− feet (bottom left) are shown, with pathways grouped into themes. Pathway details are shown in Tables S3 and S4 in the supplemental material. (B) Comparison of gene signatures of chikungunya virus (CHIKV) arthritis in WT and CCR2−/− mice with a gene signature from RA patients. Gene set enrichment analysis showed statistically significant enrichment of upregulated (UP) genes (163 genes in CCR2−/− mice and 341 genes in WT mice in “core enrichment”; overlap, 150) and downregulated (DOWN) genes (82 genes in CCR2−/− mice and 76 genes in WT mice in “core enrichment”) from CHIKV arthritis in the up- and downregulated genes, respectively, identified in synovial biopsy specimens from RA patients.