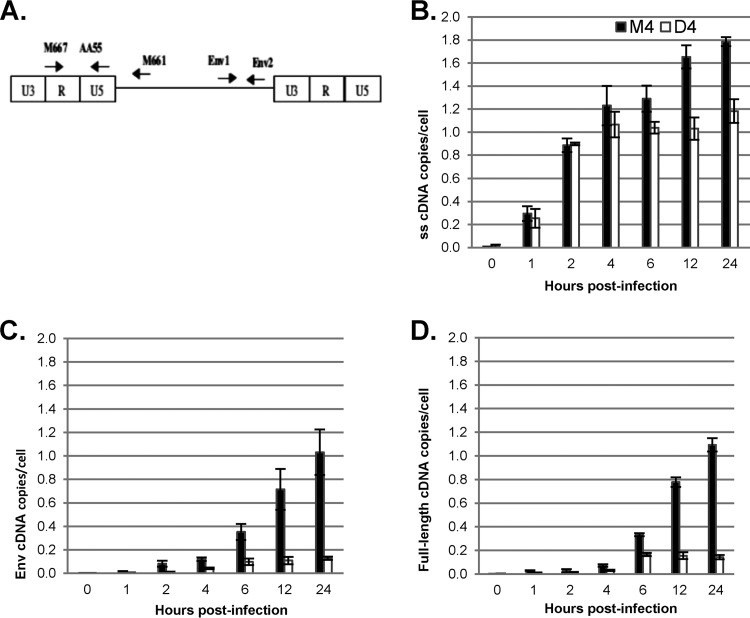
FIG 2.
DBR1 shRNA inhibits the completion of HIV-1 reverse transcription. GHOST-R5X4 cells were transfected with D4 or M4 for 48 h, followed by infection with a DNase-treated HIV-1 vector (HR-E). Zero to 24 h later, the cells were lysed and DNA was isolated for qPCR to evaluate the synthesis of HIV-1 cDNA. The number of copies of HIV-1 was normalized to the number of copies of β-globin. The data represent those from three independent time course experiments processed in triplicate. Error bars denote the standard deviations from the means. Differences between D4- and M4-treated cells were significant by Student's t test (P < 0.01) at 6, 12, and 24 h postinfection for strong-stop DNA and at all nonzero time points for intermediate and full-length cDNA. (A) Locations of primers; (B) the oligonucleotide primers M667/AA55 specific for the R/U5 region of the LTR were used to detect early reverse transcripts (strong-stop [ss] DNA); (C) env primers env1/env2 were used to detect intermediate reverse transcripts; (D) LTR/gag primers M667/M661 were chosen to detect complete, full-length viral cDNA.