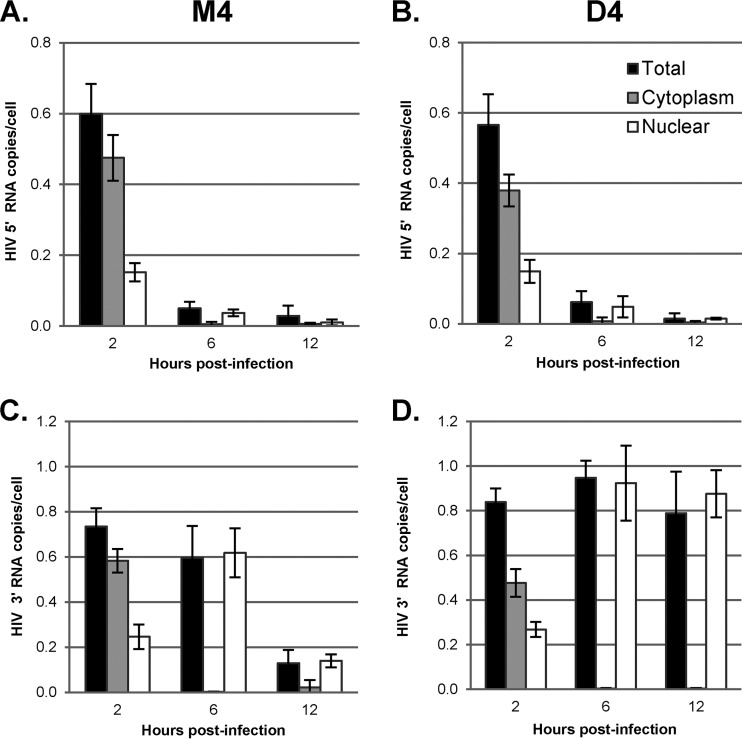
FIG 5.
Degradation and subcellular localization of the viral RNA genome during reverse transcription. GHOST-R5X4 cells were transfected with D4 or M4 for 48 h, followed by infection with a DNase-treated HIV-1 vector (HR-E). Two to 12 h later, the cells were either fractionated utilizing the NP-40 protocol or lysed to prepare whole-cell fractions. RNA was isolated for RT-qPCR to evaluate the degradation and subcellular localization of the viral RNA genome during reverse transcription. The data were normalized to the GAPDH copy number. Error bars denote the standard deviations from the means. The data represent those from three independent time course experiments processed in triplicate. LTR/gag primers M667/M661 were chosen to detect the 5′ region of the viral RNA genome in M4-treated cells (A) and D4-treated cells (B). The env primers env1/env2 were used to detect the 3′ region of the viral RNA genome in M4-treated cells (C) and D4-treated cells (D).