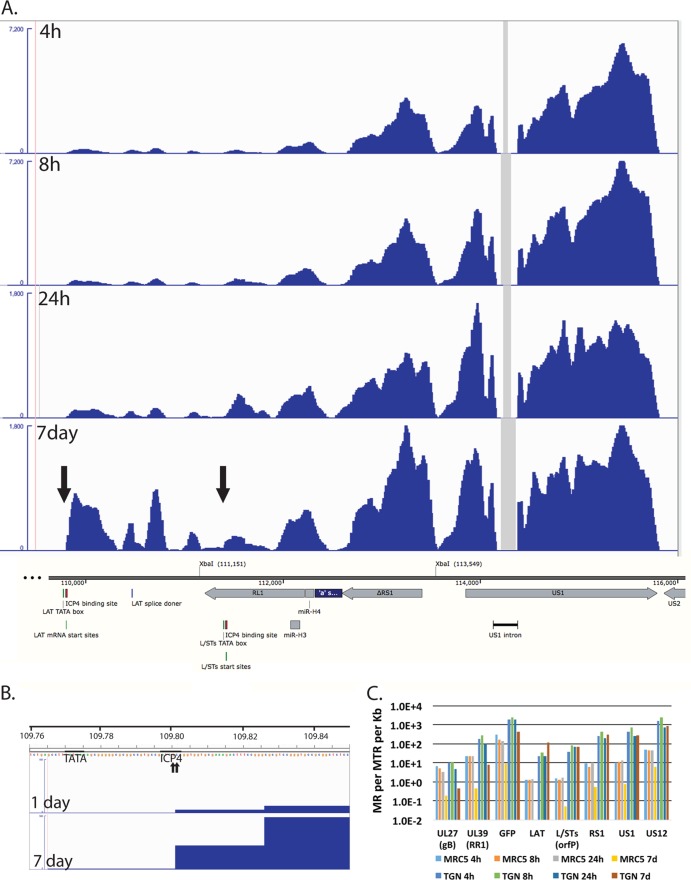
FIG 8.
Transcription of the internal repeat region from persisting genomes. (A) The RNA-seq reads from d109-infected TG neurons mapping to the region encoding the LAT promoter to US2 are shown relative to features in this region of the genome. The XbaI site at 111,151 marks the locus of the ICP0 deletion, and the site 113,549 marks the locus of the deletion that removed the VP16-responsive elements from the region. The numbers shown are in bp from the 5′ end of the modified d109 genome in the parental orientation. The maximum of the y axes for the 4- and 8-h graphs was set at 7,200 reads, while that for the 24-h and 7-day graphs was set at 1,800 reads. (B) The RNA-seq reads for the 1- and 7-day d109-infected TG neuron samples are shown in the region of the LAT TATA box, mRNA start sites, and ICP4 binding site. The maximum on the y axis is 100 reads. The beginning of the signal corresponds to the arrow to the left in the 7-day graph of panel A. (C) Quantification of the reads from the samples in Fig. 5 for select viral genes.