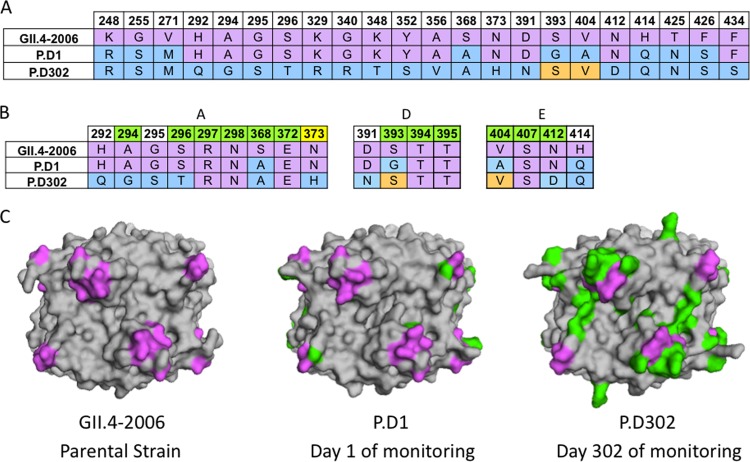
FIG 1.
Sequence changes in chronically infected patient strains compared to GII.4-2006b. (A) Available capsid amino acid sequences for GII.4-2006b, P.D1, and P.D302 were aligned using Clustal Omega, and sequence differences among GII.4-2006, P.D1, and P.D302 are shown. GII.4-2006b residues are shown in purple. P.D1 and P.D302 differences from GII.4-2006b are indicated in light blue, while orange indicates reversion to the GII.4-2006b residues. (B) Alignment of GII.4-2006b, P.D1, and P.D302 amino acid sequences in and around epitopes A, D, and E. Green indicates a position within a defined epitope, while white indicates nearby residues that may impact antigenicity in these epitopes. Yellow indicates an amino acid position newly defined as part of epitope A. (C) Structural homology models of GII.4-2006b, P.D1, and P.D302 capsid P2 dimers shown from a top view. Purple shows the locations of epitopes A, D, and E on the capsid P2 dimer, while green shows changed amino acid residues in P.D1 and P.D302 compared to GII.4-2006b.