Figure 6.
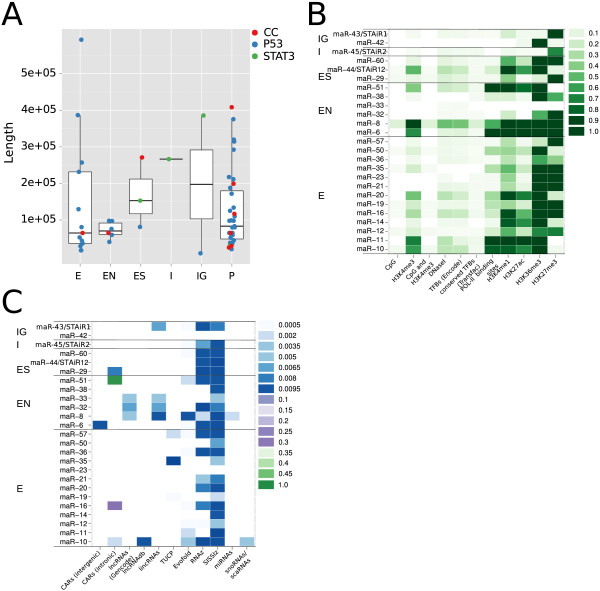
Characterization of DE-macroRNAs.(A) The size distribution of DE-macroRNAs indicates similar sizes for the different genomic categories of DE-macroRNAs (intergenic, overlapping exons, overlapping non-coding exons, located in introns, joint start but different end as coding RNA and presumed primary transcript) and throughout the three different transcriptome surveys (cell cycle, p53 and STAT3). (B) Fraction of nucleotides in DE-macroRNAs overlapping with putative promoter regions, transcription factor binding sites, polII binding sites and epigenetically modified regions. (C) Fraction of nucleotides in DE-macroRNAs overlapping with known ncRNA annotations. Annotations are described in detail in Additional file 1: Table S28. CC, cell cycle; E, overlapping exons; EN, overlapping non-coding exons; ES, joint start but different end as coding RNA; I, located in introns; IG, intergenic; P, presumed primary transcript; STAT3, signal transducer and activator of transcription-3.
