Figure 4.
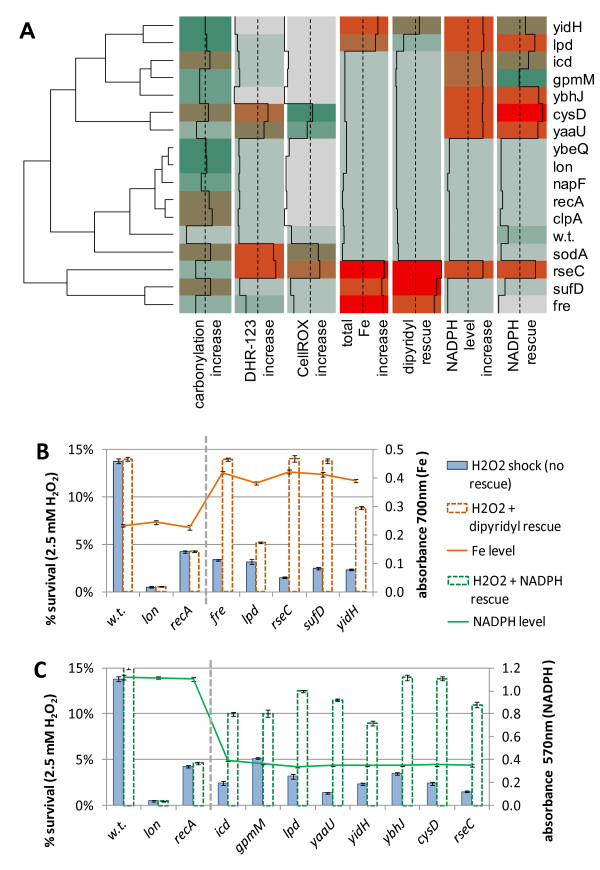
Mechanism of activity of the putative oxidative stress protection genes. (A) Overview of outcomes of seven experimental assays (columns) performed with the wild-type Escherichia coli and 15 deletion mutants. A larger value denotes a stronger observed effect; values are adjusted so that 0 signifies no effect and 1 signifies strong effect (values <0 and >1 are possible; for details of normalization for each assay, see Additional file 1). DHR-123 and CellROX are fluorescent dyes that measure reactive oxygen species. 2,2′-dipyridyl is an iron chelator. Genes are clustered based on similarity of the normalized response profiles of the mutants across the assays. Dashed lines denote the median. (B, C) A detailed display of the non-normalized measurements of: (B) iron levels and survival in the dipyridyl rescue experiment, or (C) NADPH levels and survival in the corresponding rescue experiment. Data are shown for wild-type E. coli, for lon and recA mutants (well-investigated genes expected not to act by the examined mechanisms), and those candidate genes in which our experiments support the proposed mechanism of action. Error bars show the 95% CI of the mean, determined over at least four replicates.
