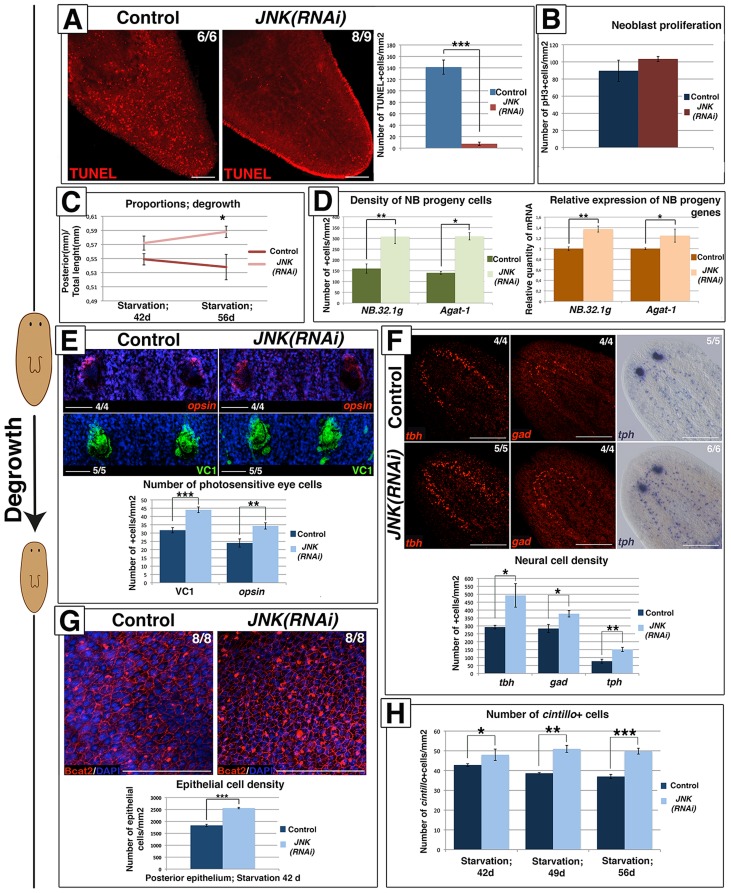
Figure 6. JNK-dependent apoptosis is required for proper remodeling during degrowth.
(A) Whole-mount TUNEL staining showing apoptotic cell death during degrowth in post-pharyngeal regions and graph showing quantification of cells dying by apoptosis (TUNEL-positive) in animals starved for 42 days. At least six biological replicates were used. (B) Graph showing quantification of mitotic (pH3+) cells in animals starved for 42 days. At least nine biological replicates were used. (C) Graph showing the size of the post-pharyngeal region (from pharynx anchoring to tail tip) relative to the whole-body length during degrowth. Values represent the means of 15 biological replicates. (D) Quantification of NB.32.1g and Agat-1 expression in starved animals. The green histograms depict the quantification of cells positive for these two markers after FISH. At least four biological replicates were used. The orange histograms depict the relative expression of the two markers as determined by qRT-PCR. Values represent the means of three biological replicates. (E). FISH analysis of photoreceptor cells (opsin+), immunostaining of the visual system (VC1+) with DAPI counterstaining and graph showing the quantification of the number of photoreceptor cells/eye in animals starved for 42 days. Four biological replicates (8 eyes) were used. (F) FISH analysis of GABAergic (gad+) and octopaminergic (tbh+) neurons, WISH analysis of serotoninergic (tph+) neurons and graph showing the quantification of the number of neural cells/mm2 in the brains of animals starved for 42 days. (G) Staining of the epithelia with DAPI and anti-β-catenin-2 antibody (Bcat2), and graph showing the quantification of the number of epithelial cells/mm2 in post-pharyngeal regions of animals starved for 42 days. Eight biological replicates were used. (H) Quantification of the number of cintillo+ cells in the anterior region during the starvation process. At least five biological replicates were used. (Top left, anterior). All images, except for the tph image, correspond to confocal z-projections. Error bars represent standard error of the mean. Data were analyzed by Student's t-test. *P<0.05; **P<0.01; ***P<0.001; differences are considered significant at P<0.05. Scale bars: 100 µm (A), 50 µm (E), 300 µm (F), 50 µm (G). d, days of starvation.