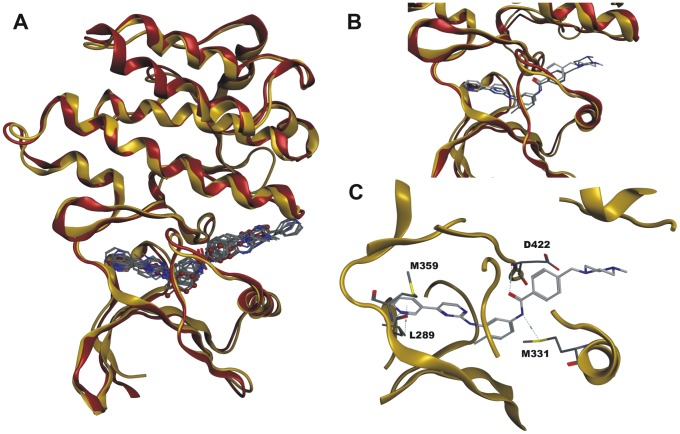
Figure 1. Homology models of human and schistosome Abl kinases.
Panels A to C: Structural comparisons of the molecular docking solutions of the SmAbl2 homology model versus the human Abl2 crystal structure (3GVU). Both protein structures are shown in ribbon representation, the SmAbl2 homology model is colored yellow, while the human Abl2 crystal structure is colored red. A: Structural superposition of the two protein structures. The ten highest scoring docking solutions for Imatinib in the SmAbl2 homology model were depicted in stick representation in atom color, the crystal structure pose of Imatinib in the human Abl2 crystal structure is depicted in ball-and-stick representation colored red. Hydrogen atoms were omitted for visibility reasons in the depiction. The RMSD value of the template and the model structure resulted in a Cα RMSD (root-mean-square deviation) value of 0.765 Å of the corresponding 245 amino acid residues. B: Close-up view of the binding sites of the two protein structures. The top-scored pose of Imatinib in the SmAbl2 homology model was depicted in stick representation with light-grey carbon atoms, the crystal structure pose of Imatinib in human Abl2 was depicted in stick representation with dark grey carbon atoms. C: Close-up view of the binding site of SmAbl2 homology model. Imatinib engages in four directed interactions to surrounding amino acid residues, depicted by light blue dashed lines.