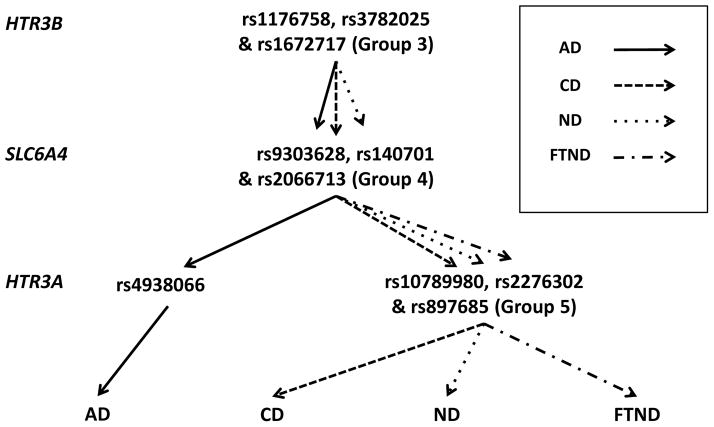
Figure 7.
Summary of detected interaction models in the EA sample. GMDR-GPU (Zhu et al. 2013) was used to perform exhaustive searches for two- to five-way interaction models. The best interaction models for AD, CD, ND, and FTND shown in the figure were determined based on CVC >7 of 10 and prediction accuracy > 55%. The P value associated for each model shown here was < 0.005 based on 107 permutation tests. Interaction models with different phenotypes involved overlapped and highly correlated SNPs, which were grouped together. Group 3 includes rs1176758, rs3782025 and rs1672717; Group 4 includes rs9303628, rs140701 and rs2066713; Group 5 includes rs10789980, rs2276302 and rs897685. Pair-wise D values of adjacent SNPs within each group are greater than 0.7. SNP combinations are color-coded.