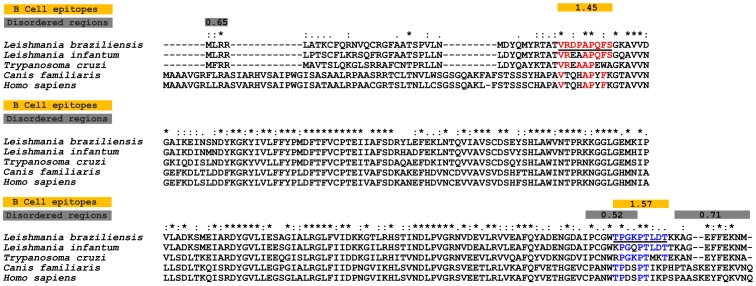
Figure 1. Sequence divergence and prediction of B-cell linear epitopes and intrinsically unstructured/disordered regions in L. braziliensis Peroxidoxin and its orthologs.
Alignment between L. braziliensis peroxidoxin (TritrypDB ID: LbrM.23.0050) and orthologous proteins present in L. infantum (TritrypDB ID: Linj.23.0050), T. cruzi (TritrypDB ID:TcCLB.509499.14), H. sapiens (RefSeq ID: NP_006784.1) and C. familiaris (RefSeq ID: NP_001243414.1). The box signifies conserved domains present in the peroxidoxin protein of L. braziliensis, as identified by CDD NCBI [48]. The yellow boxes mark predicted B-cell epitopes, and the gray boxes mark predicted disordered regions. The continuous black underlined amino acid sequences represent two potential B-cell epitopes predicted by Bepipred in the LbrM.23.0050 protein, and the colors highlight amino acid conservations in the T. cruzi, C. familiaris and H. sapiens sequences in relation to the L. braziliensis sequence.