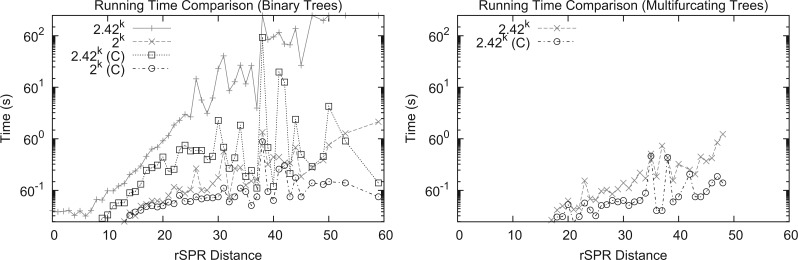
Figure 4.
Mean time required to compare gene trees with a given SPR distance from an SPR supertree of a 244-genome data set. The time axis is on a log scale as the time required increases exponentially with the SPR distance. The left panel compares our previous (2.42kn) and new (2kn) algorithms, with (C) and without clustering, on the set of binary trees. The right panel compares our new algorithm with and without clustering on the set of trees with unsupported bipartitions collapsed. Note that collapsing bipartitions reduces the SPR distance.