Abstract
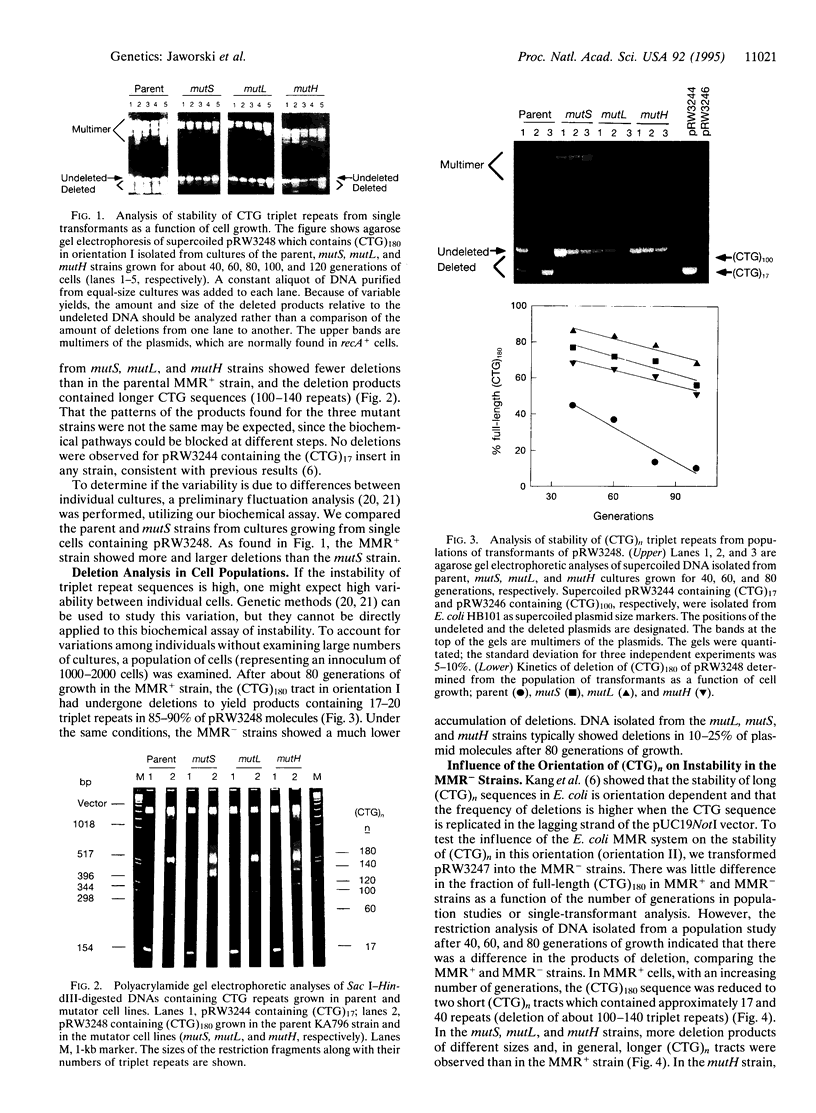
Long CTG triplet repeats which are associated with several human hereditary neuromuscular disease genes are stabilized in ColE1-derived plasmids in Escherichia coli containing mutations in the methyl-directed mismatch repair genes (mutS, mutL, or mutH). When plasmids containing (CTG)180 were grown for about 100 generations in mutS, mutL, or mutH strains, 60-85% of the plasmids contained a full-length repeat, whereas in the parent strain only about 20% of the plasmids contained the full-length repeat. The deletions occur only in the (CTG)180 insert, not in DNA flanking the repeat. While many products of the deletions are heterogeneous in length, preferential deletion products of about 140, 100, 60, and 20 repeats were observed. We propose that the E. coli mismatch repair proteins recognize three-base loops formed during replication and then generate long single-stranded gaps where stable hairpin structures may form which can be bypassed by DNA polymerase during the resynthesis of duplex DNA. Similar studies were conducted with plasmids containing CGG repeats; no stabilization of these triplets was found in the mismatch repair mutants. Since prokaryotic and human mismatch repair proteins are similar, and since several carcinoma cell lines which are defective in mismatch repair show instability of simple DNA microsatellites, these mechanistic investigations in a bacterial cell may provide insights into the molecular basis for some human genetic diseases.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aquilina G., Hess P., Branch P., MacGeoch C., Casciano I., Karran P., Bignami M. A mismatch recognition defect in colon carcinoma confers DNA microsatellite instability and a mutator phenotype. Proc Natl Acad Sci U S A. 1994 Sep 13;91(19):8905–8909. doi: 10.1073/pnas.91.19.8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backman K., Betlach M., Boyer H. W., Yanofsky S. Genetic and physical studies on the replication of ColE1-type plasmids. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):69–76. doi: 10.1101/sqb.1979.043.01.012. [DOI] [PubMed] [Google Scholar]
- Cleaver J. E. It was a very good year for DNA repair. Cell. 1994 Jan 14;76(1):1–4. doi: 10.1016/0092-8674(94)90165-1. [DOI] [PubMed] [Google Scholar]
- Eichler E. E., Holden J. J., Popovich B. W., Reiss A. L., Snow K., Thibodeau S. N., Richards C. S., Ward P. A., Nelson D. L. Length of uninterrupted CGG repeats determines instability in the FMR1 gene. Nat Genet. 1994 Sep;8(1):88–94. doi: 10.1038/ng0994-88. [DOI] [PubMed] [Google Scholar]
- Fishel R., Ewel A., Lee S., Lescoe M. K., Griffith J. Binding of mismatched microsatellite DNA sequences by the human MSH2 protein. Science. 1994 Nov 25;266(5189):1403–1405. doi: 10.1126/science.7973733. [DOI] [PubMed] [Google Scholar]
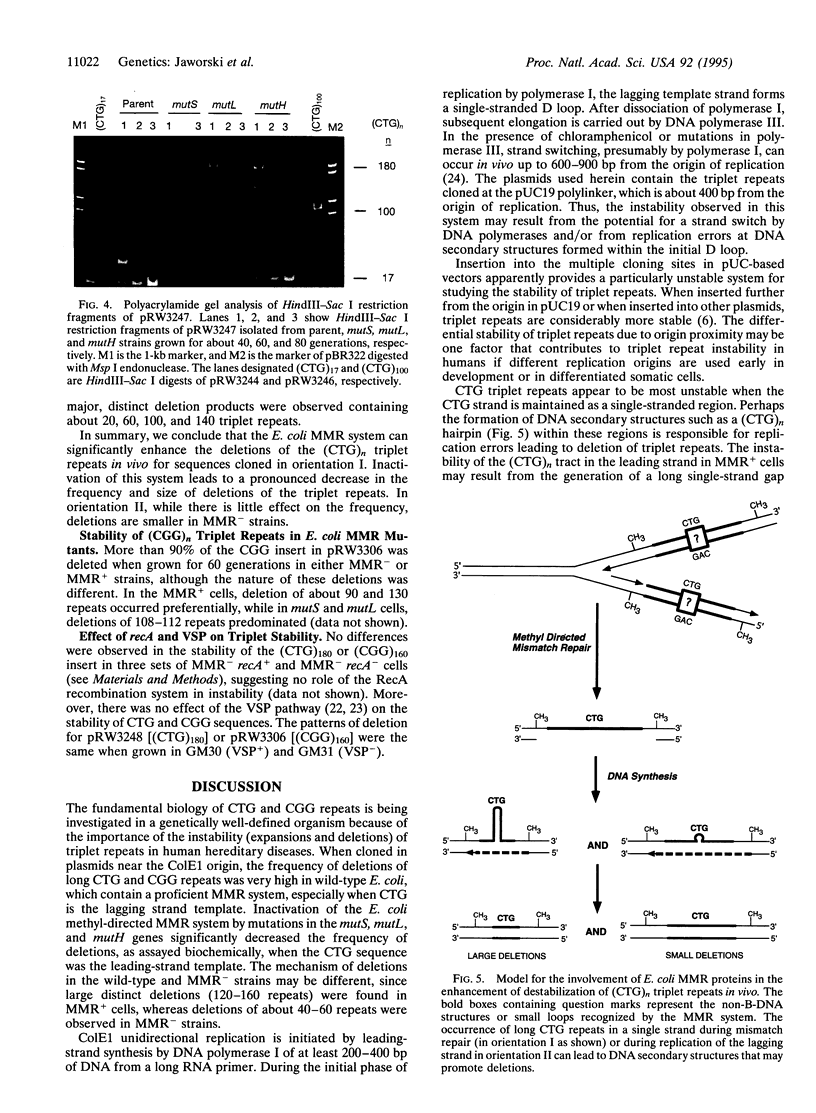
- Kang S., Jaworski A., Ohshima K., Wells R. D. Expansion and deletion of CTG repeats from human disease genes are determined by the direction of replication in E. coli. Nat Genet. 1995 Jun;10(2):213–218. doi: 10.1038/ng0695-213. [DOI] [PubMed] [Google Scholar]
- Karran P., Bignami M. DNA damage tolerance, mismatch repair and genome instability. Bioessays. 1994 Nov;16(11):833–839. doi: 10.1002/bies.950161110. [DOI] [PubMed] [Google Scholar]
- Kolodner R. D., Alani E. Mismatch repair and cancer susceptibility. Curr Opin Biotechnol. 1994 Dec;5(6):585–594. doi: 10.1016/0958-1669(94)90079-5. [DOI] [PubMed] [Google Scholar]
- Levinson G., Gutman G. A. High frequencies of short frameshifts in poly-CA/TG tandem repeats borne by bacteriophage M13 in Escherichia coli K-12. Nucleic Acids Res. 1987 Jul 10;15(13):5323–5338. doi: 10.1093/nar/15.13.5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb M., Rehmat S. Very short patch repair of T:G mismatches in vivo: importance of context and accessory proteins. J Bacteriol. 1995 Feb;177(3):660–666. doi: 10.1128/jb.177.3.660-666.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luria S. E., Delbrück M. Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics. 1943 Nov;28(6):491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus M. G. Location of DNA methylation genes on the Escherichia coli K-12 genetic map. Mol Gen Genet. 1973 Dec 14;127(1):47–55. doi: 10.1007/BF00267782. [DOI] [PubMed] [Google Scholar]
- Modrich P. Mechanisms and biological effects of mismatch repair. Annu Rev Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- Modrich P. Mismatch repair, genetic stability, and cancer. Science. 1994 Dec 23;266(5193):1959–1960. doi: 10.1126/science.7801122. [DOI] [PubMed] [Google Scholar]
- Nicolaides N. C., Papadopoulos N., Liu B., Wei Y. F., Carter K. C., Ruben S. M., Rosen C. A., Haseltine W. A., Fleischmann R. D., Fraser C. M. Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature. 1994 Sep 1;371(6492):75–80. doi: 10.1038/371075a0. [DOI] [PubMed] [Google Scholar]
- Parker B. O., Marinus M. G. Repair of DNA heteroduplexes containing small heterologous sequences in Escherichia coli. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1730–1734. doi: 10.1073/pnas.89.5.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons R., Li G. M., Longley M. J., Fang W. H., Papadopoulos N., Jen J., de la Chapelle A., Kinzler K. W., Vogelstein B., Modrich P. Hypermutability and mismatch repair deficiency in RER+ tumor cells. Cell. 1993 Dec 17;75(6):1227–1236. doi: 10.1016/0092-8674(93)90331-j. [DOI] [PubMed] [Google Scholar]
- Richards R. I., Sutherland G. R. Simple repeat DNA is not replicated simply. Nat Genet. 1994 Feb;6(2):114–116. doi: 10.1038/ng0294-114. [DOI] [PubMed] [Google Scholar]
- Shibata D., Peinado M. A., Ionov Y., Malkhosyan S., Perucho M. Genomic instability in repeated sequences is an early somatic event in colorectal tumorigenesis that persists after transformation. Nat Genet. 1994 Mar;6(3):273–281. doi: 10.1038/ng0394-273. [DOI] [PubMed] [Google Scholar]
- Strand M., Prolla T. A., Liskay R. M., Petes T. D. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature. 1993 Sep 16;365(6443):274–276. doi: 10.1038/365274a0. [DOI] [PubMed] [Google Scholar]
- Umar A., Boyer J. C., Kunkel T. A. DNA loop repair by human cell extracts. Science. 1994 Nov 4;266(5186):814–816. doi: 10.1126/science.7973637. [DOI] [PubMed] [Google Scholar]
- Umar A., Boyer J. C., Thomas D. C., Nguyen D. C., Risinger J. I., Boyd J., Ionov Y., Perucho M., Kunkel T. A. Defective mismatch repair in extracts of colorectal and endometrial cancer cell lines exhibiting microsatellite instability. J Biol Chem. 1994 May 20;269(20):14367–14370. [PubMed] [Google Scholar]
- Willems P. J. Dynamic mutations hit double figures. Nat Genet. 1994 Nov;8(3):213–215. doi: 10.1038/ng1194-213. [DOI] [PubMed] [Google Scholar]