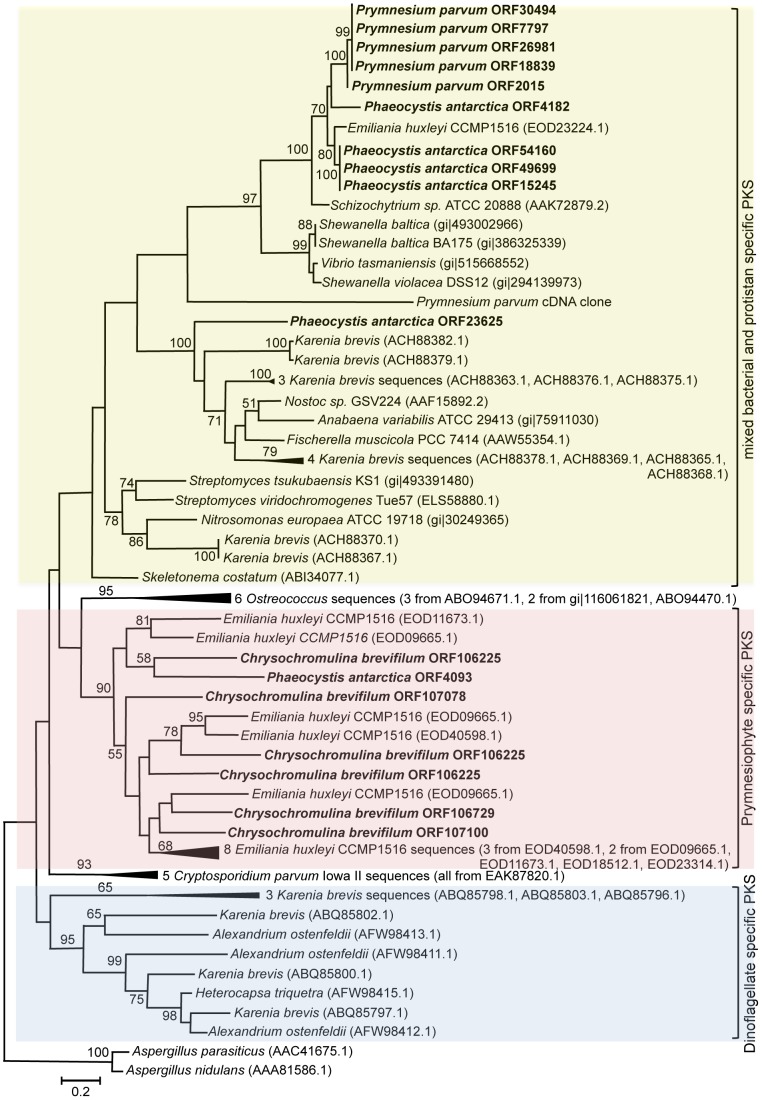
Figure 2. Polyketide synthase maximum likelihood tree with 100 iterated bootstraps using only the keto-synthase (KS) domain.
The tree was inferred using MEGA5 (Tamura et al. 2011) with maximum likelihood method based on Jones-Taylor-Thornton model. The analysis involved 78 amino acid sequences. All positions with less than 95% site coverage were eliminated. There were 181 total sites in the final dataset. Bootstrap support values, if greater than 50%, are shown as the percentages of 100 trees inferred in the analysis. The scale bar represents the number of substitutions per site. The tree is rooted with Aspergillus nidulans polyketide synthase. Sequences from our dataset are shown in bold. Multiple branches have the same identifying ORFs,GI or accession numbers due to multiple KS domains on the same gene.