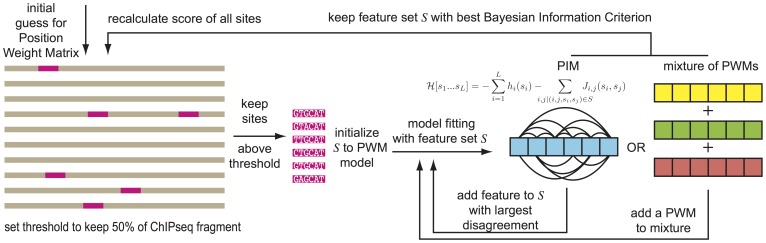
Figure 1. Workflow.
An initial Position Weight Matrix (PWM) is used to find a set of binding sites on ChIPseq data. Models are then learned using single-point frequencies (PWM), two-point correlations (PIM) or a mixture of PWM models learned on sites clustered by K-Means with increasing complexity, i.e. increasing number of features in the model. Finally the models with best Bayesian Information Criteria (BIC) are used to predict new binding sites until convergence to a stable set of TFBSs.