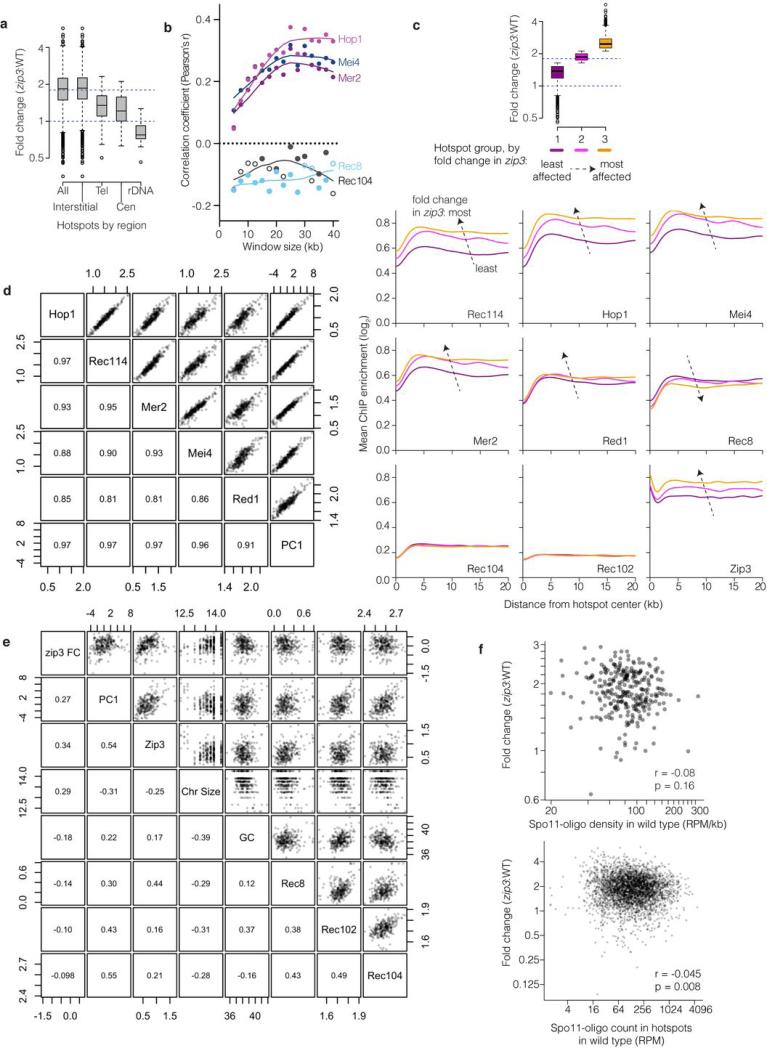
Extended Data Figure 9. Changes in the DSB landscape in zip3.
a, Change in Spo11-oligo counts in hotspots grouped by chromosomal context. Tel, within 20 kb of telomeres; Cen, within ±10 kb of centromeres; rDNA, from 60 kb leftward to 30 kb rightward of rDNA; Interstitial, all others. Dashed lines mark values assumed as no change and average change (1.8-fold). Boxes indicate median and interquartile range; whiskers indicate the most extreme data points which are ≤1.5 times the interquartile range from the box; individual points are outliers. Sub-telomeric and pericentric zones show less increase in zip3 on average, thus, ZMM-dependent feedback contributes less than other, unknown factors to suppressing DSBs in these regions. The zone near the rDNA showed no increase or was even decreased; thus, zip3 mutants are competent for this region's DSB suppression, which is dependent on the ATPase Pch2 and the replication factor Orc1 (ref 63). Note that the remaining interstitial hotspots showed highly variable response to zip3 mutation (>20 fold). b, Correlation between log-fold change in Spo11-oligo counts in zip3 and the binding of the indicated proteins, binned in non-overlapping windows of varying size. Closed symbols, p<0.05. ChIP data are from ref 38. c, Average ChIP profiles around interstitial hotspots divided into three equal-sized groups according to the average fold change in zip3. Top: the box-and-whisker plot (as described for panel a) shows the distribution of fold changes for the three groups. Below: ChIP profiles for each of the indicated proteins. Note that the profiles lie atop one another for Rec102 and Rec104. Dashed arrows indicate direction of the change in the average profiles with increasing fold change in zip3. ChIP data are from refs 38 and 39. d, High degree of colinearity of log2-transformed ChIP data38 for Rec114, Mei4, and Mer2 (which are essential for DSB formation) and Hop1 and Red1 (axis proteins that promote normal DSB formation). More than 90% of the variance for this combination of ChIP data is captured in the first principal component (PC1). The high degree of correlation between these proteins was described previously38. e, Correlations between the fold change in zip3 (zip3 FC, log2 and assuming 1.8-fold increase genome-wide) and various chromosomal features: principal component 1 (PC1) for Rec114, Mei4, Mer2, Hop1, and Red1 ChIP data (same as in panel d); chromosome size (loge(bp)); G+C content (%); and ChIP data for the indicated proteins (log2). In d and e, upper right panels show pair-wise scatter plots and lower left panels show corresponding correlation coefficients (Pearson's r) for data for interstitial regions binned in 35-kb non-overlapping windows. Essentially identical results were obtained with different window sizes (20–40 kb) or with varying placement of windows (data not shown). f, Essentially no correlation between DSB activity in wild type and change in zip3, whether considering interstitial regions divided into non-overlapping 35-kb bins (upper panel) or interstitial hotspots (lower panel). A 1.8-fold increase genome-wide in zip3 is assumed. Note: Fold change is labeled according to a linear scale but plotted in a log scale in panels a, c, f.