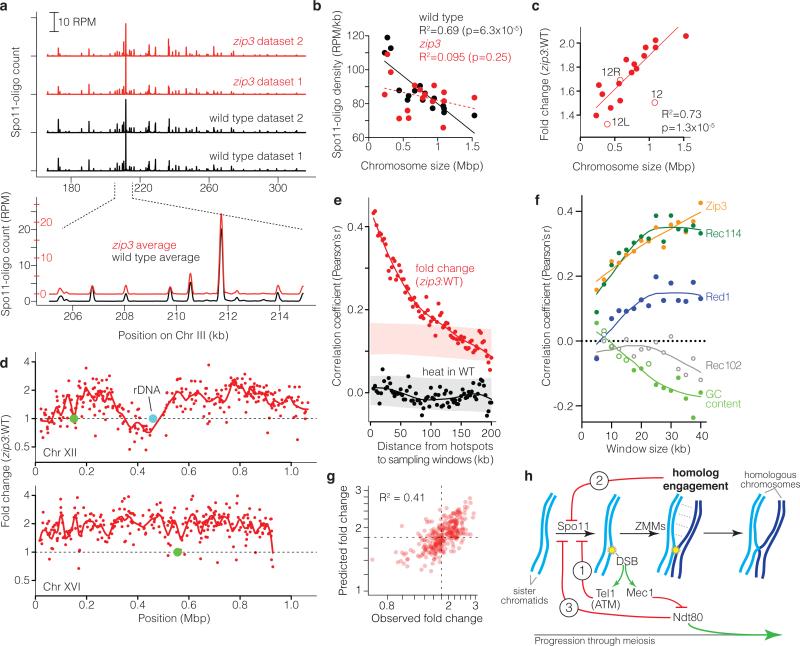
Figure 4. Altered DSB distribution in zip3 mutants.
a, Upper, reproducibility of Spo11-oligo maps. Lower, DSBs form at the same hotspots in zip3 as wild type. Smoothed with 201-bp Hann window. b, Zip3 is required for chromosome size-dependent variation in Spo11-oligo density. Lines, least squares fits (dashed = not significant). c, Larger chromosomes experience greater increase in Spo11 oligos. Fold change is the per-chromosome Spo11-oligo density in zip3 over wild type (WT). Open circles, Chr XII (“12”, omitting rDNA length) and the portions of Chr XII left or right of the rDNA (“12L”, “12R”). Regression line treats 12L and 12R as separate chromosomes. d, Regional variation in response to zip3 mutation. Each point is the change at a hotspot (plotted on log scale). Red lines, local regression (loess); green circles, centromeres. e, Local domains of correlated behavior. Each point compares hotspots to their neighbors in 5-kb-wide windows the indicated distance away. Nearby hotspots show correlated behavior for fold change in zip3 (red), but not heat (Spo11-oligo frequency) in wild type (black). Shaded areas denote 95% CI estimates for hotspots randomized within-chromosome (randomized r>0 for zip3-fold-change because of the chromosome size effect). f, Correlation between log-fold-change in zip3 and binding of indicated proteins, binned in non-overlapping windows of varying size. For clarity, other proteins are in Extended Data Fig. 9b. Pericentric, sub-telomeric, and rDNA-proximal regions were censored. Closed symbols, p<0.05. g, Fit of multiple regression model predicting changes (log scale) in Spo11-oligo density in 35-kb windows from ChIP data, G+C content, and chromosome size (Supplementary Table 5). Dashed lines, observed mean fold change. h, Network of feedback circuits controlling DSB formation. Circuit 1: DSBs activate Tel1 (ATM in mouse), which inhibits further DSB formation. Circuit 2: ZMM-dependent interactions between homologous chromosomes inhibit Spo11. Circuit 3: Ndt80 shuts down DSB formation and drives pachytene exit; Mec1 kinase delays or blocks Ndt80 activation when DSBs are present.