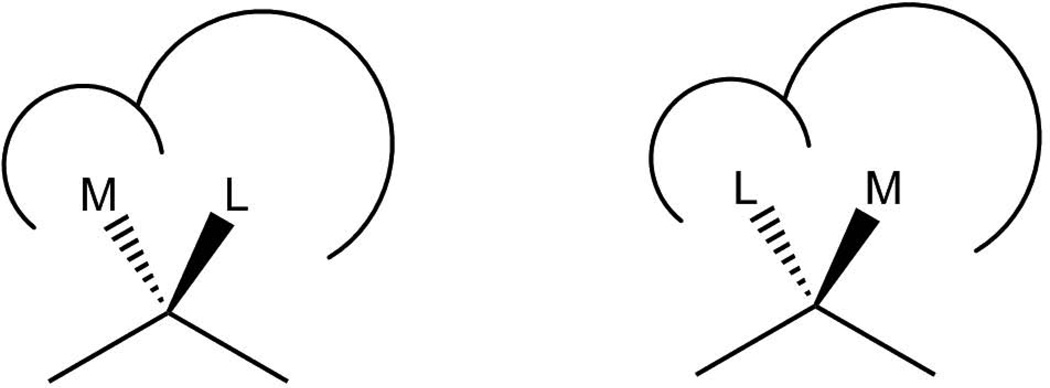Abstract
The prospect for computer aided refinement of stereoselective enzymes is further validated by simulating the ester hydrolysis by the wild type and mutants of CalB, focusing on the challenge of dealing with strong steric effects and entropic contributions. This was done using the empirical valence bond (EVB) method in a quantitative screening of the enantioselectivity, considering both kcat and kcat/KM of the R and S stereoisomers. Although the simulations require very extensive sampling for convergence they give encouraging results and major validation, indicating that our approach offers a powerful tool for computer aided design of enantioselective enzymes. This is particularly true in cases with large changes in steric effects where alternative approaches may have difficulties in capturing the interplay between steric clashes with the reacting substrate and protein flexibility.
Keywords: EVB, free energy calculations, serine hydrolase, mutation, stereoselective enzyme, enzyme design, transesterification
I. Introduction
Optimizing enzymes to catalyze selective enantioselective reactions has a major potential in biotechnology.1 For example, the use of biocatalysts for efficient synthesis of enantiomerically pure chiral molecules is of great importance in the production of drugs by the pharmaceutical industry.2 Unfortunately, quantifying the observed enantioselectivity in different enzymes presents a significant challenge for approaches aimed at understanding enzyme catalysis. Experimental studies of enantioselective enzymatic reactions have provided major advances in recent years (e.g., ref3–5). The main focus of these studies turned to lipases3,6–13 via an examination of esterification reactions14, solvent effects15, the temperature effects7 and substrate effects8–10. Furthermore, instructive advances have been done with directed evolution experiments.3,11
Enantioselective enzymatic reactions have also been examined by theoretical approaches, including MM, MD studies9,13,14,16 and QM/MM17, but these interesting studies have not provided quantitative insight. Attempts to use the cluster QM model18,19 have provided interesting insight and encouraging results but such an approach might find difficulties in capturing entropic effects and in overestimating strong steric effects.
Overall it is important to explore QM/MM approaches that involve extensive sampling and evaluate the actual activation free energies, since such strategies should be able to explore entropic effects and to allow for exploring more realistic relaxation of the active site.
In trying to obtain more quantitative results it is crucial to improve two aspects of the modeling; namely the potential surface and the sampling. That is, trying to evaluate the free energy of mutating the R to S enantiomers using a force field model of their TS can be useful, but here it is important to determine the correct charge distributions and structure of the TS and this can be best done by a QM/MM approach (it is important to capture the change of the TS charge and geometry upon interaction with the entire enzyme while considering its flexibility consistently). Unfortunately, the use of a QM/MM approach is unlikely to give reliable free energies without extensive sampling (in fact, the sampling of the enzyme substrate configurations is also the most crucial requirement in classical force field studies). Here, the empirical valence bond (EVB)20,21 arguably provides the optimal current strategy, since it combines a reliable semiempirical QM/MM model with the ability for extremely effective sampling. In fact a previous study of enantioselectivity of Candida Antarctica lipase A (CalA) by the EVB approach22 provided encouraging results. However, further general validation is still needed. Here we focus on the catalysis of the enzyme Candida antarctica lipase B (CalB)23 which is used heavily in industrial organic synthesis by exhibiting unprecedented enantioselectivity towards (R)-phenylethanole7. Interestingly, this peculiar behavior is being reversed to favor (S)-phenylethanole upon mutating the active site tryptophan into the smaller alanine residue7. More specifically ,the apparent kinetic rate constant and the apparent Michaelis constant, i.e. and , for the wild-type (WT) CalB and the W104A mutation, were determined for the acylation of S-phenylethanole with vinyl-butanoate in cyclohexane at 30°C. The experimental results indicate a change in enantioselectivity by a factor of 8300000 through the point-mutation in the stereoselectivity pocket, which was mainly achieved by increasing towards the slow-reacting S-configured enantiomer. More specifically, towards S-phenylethanole was 64000 times larger for the mutant compared to WT CalB, while the same constant decreased by a factor of 130 toward the R-enantiomer. Interestingly, remained basically unchanged for the W104A mutation and was almost equal towards the R and S enantiomers, while they both decreased by a factor of two compared to the wild type. Hence, the catalysis lies equally in kcat and KM, and the corresponding activation barriers are given in Table I. These particular experimental results for the WT are of great interest for computer simulations, as the experimentally measurable quantities in such a highly enantioselective systems clearly meet any sensitivity that may be obtained by experimental techniques, especially for the slow-reacting enantiomer, while computer simulation do, in fact, not suffer from this limitation hence may support and refine the experiment.
Table I.
The observed activation free energies for WT CalB and its W104A mutant (MUT) (a)
| Type-Substrate | Δgcat,exp |
|---|---|
| WT-R | 13.8 |
| WT-S | 22.1 |
| MUT-R | 16.7 |
| MUT-S | 15.5 |
Energies are given in kcal/mol obtained from the observed rate constants1 using transition state theory.
Further experimental analysis explored the entropic and enthalpic contributions to the enantioselectivity in the W104A mutant7. Very large entropic contributions (−T ΔS of 5.0, 8.1 and 8.4 kcal/mol in CH3CN, Decaline and Cyclohexane) were found, indicating that the origin of the selectivity can be quite complex.
The rationale for the above change in enantioselectivity has been related to the extension of the size of the so-called stereoselectivity pocket24 (as will be discussed in detail in section IV.2 and IV.4), leading to the high enantioselectivity for the R-enantiomer seen for the WT, while the W104A mutant shows a moderate selectivity for the S-enantiomer. However, determining the actual origin of the free energy changes that are involved in the steric effect is a major challenge. A similar challenge is associated with the analysis of the observed entropic contribution. These computational and fundamental challenges are met in the present work.
II. Background
II.1 The Catalytic Reaction
CalB is a serine hydrolase25,26 whose catalytic mechanism has been studied extensively (for e.g.,2,6–10,13–17,24,27–39). The CalB active site includes the catalytic triad of SER-HIS-ASP, which acts in the same way as the well-studied serine proteases21,40,41 where a proton transfer from serine is followed by a nucleophilic attack of the ester carbonyl by the deprotonated alcohol (see Figure 1). The enzyme catalyzes the reaction by stabilizing the negatively charged oxyanion by an oxyanion hole (the same type of oxyanion hole is a key catalytic factor in various proteases40,41) and by the electrostatic interaction between Asp187 and the ionized His224 (again in analogy with serine proteases21,42).
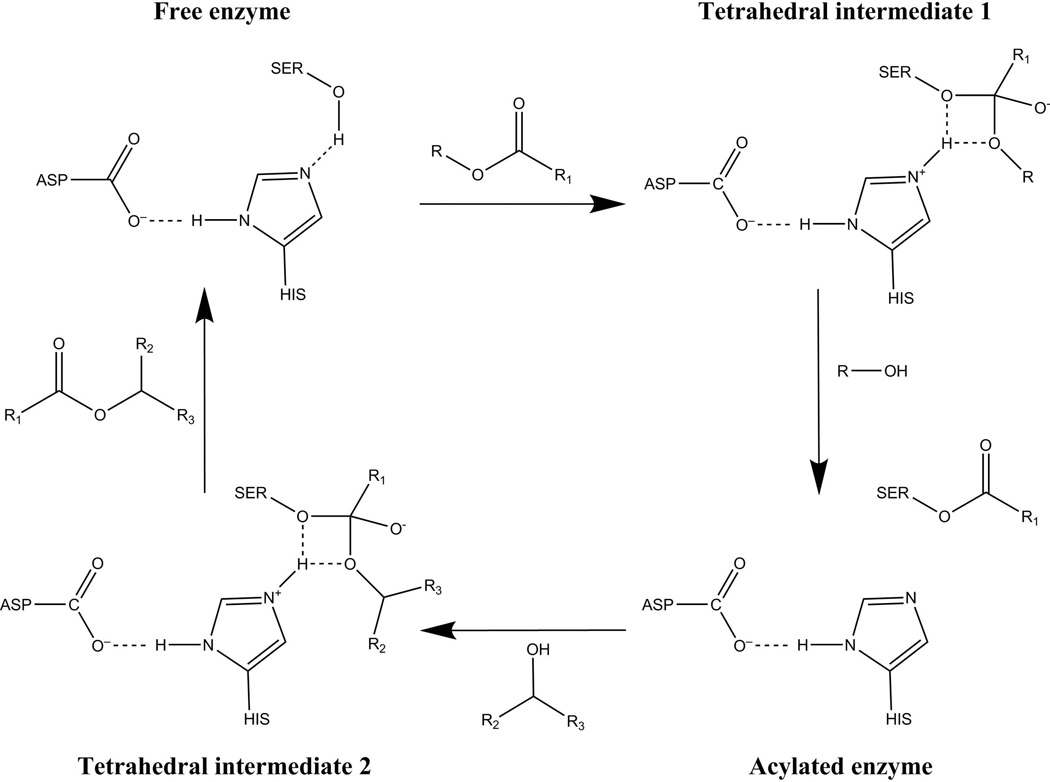
Figure 1.
A schematic of the acylation/deacylation reaction catalyzed by CalB. The reaction is initiated by the acylation step – “Free enzyme” to “Acylated enzyme” in the scheme – leading to the acylation of the active-site serine residue. The subsequent de-acylation - from the previously obtained “Acylated enzyme” to “Free enzyme” in the scheme – is established by the secondary alcohol substrate which finally results in the release of the esterified alcohol, while the “Free enzyme” in the scheme is being recovered.
The reaction can be rate limiting by either the acylation or the deacylation steps7,11 and in the case of transesterifications of secondary alcohols by CalB studied here, the deacylation step is rate limiting.11,32 That is, after the serine has been acylated (the steps leading to the “acylated enzyme” in Figure 1), the reaction proceeds via a proton transfer from the secondary alcohol to the HIS residue, followed by the rate-limiting nucleophilic attack of the acylated-seryl ester leading to the observed transesterification - the production of the chiral alcohol protected by the acyl-group (the steps leading to the esterified secondary alcohol and the recovery of “Free enzyme” in Figure 1).
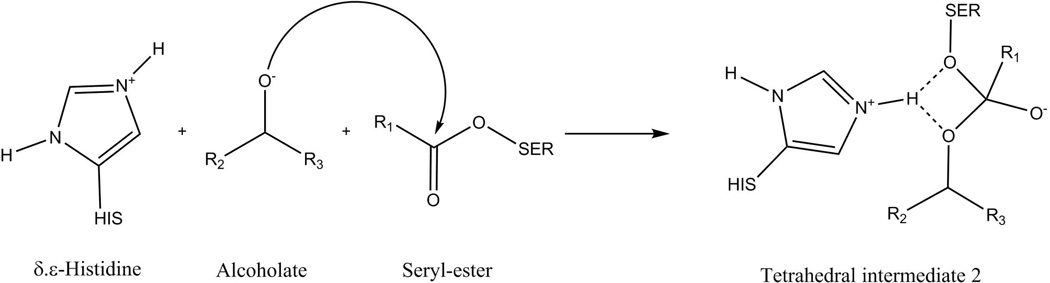
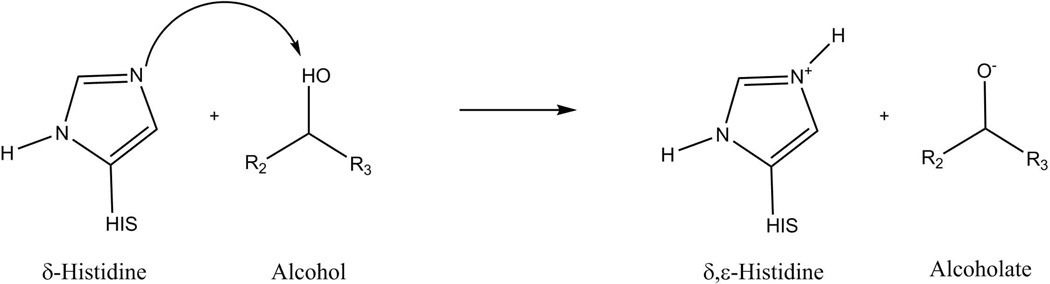
While there is controversy on whether the deacylation in this reaction may proceed via a stepwise or a concerted mechanism, the catalytic effects in both mechanisms are expected to be very similar.43 However, the stepwise calculations require the reaction to be distilled into a proton transfer step as well as a nucleophilic attack. These are given in detail in Figures 2 and 3 for the proton transfer and the nucleophilic attack, respectively, and simulation details are given in section III.
Figure 2.
Describing the proton abstraction by the imidazole base as part of the stepwise mechanism. The resulting intermediate products are doubly protonated HIS, i.e.δ,ε-Histidine, and the alcoholate nucleophile.
Figure 3.
Describing the nucleophilic attack of the secondary alcoholate as part of the stepwise mechanism. The breakdown of the product “Tetrahedral intermediate 2” yields the desired acylated secondary alcohol.
II.2 Defining the Selectivity
Here we explore the enantioselectivity (E), defined by:
| (1) |
where the notation “fast” stands for the enantiomer with the larger kcat/KM. The free energies that are relevant to kcat/KM (or more precisely to ) can be expressed in terms of the TS binding free energy21,44:
| (2) |
where is the activation barrier that corresponds to (see ref44).
As seen from Eq. 2, it is enough to mutate the R to S in the TS in the protein and then just to subtract the corresponding results for the mutation in water (which can be different than zero due to force field artifacts). The approach of mutating the TS has already been used in our early mutational studies45, but at that time we mutate the protein, while here we mutate the substrate.
In the present case there is almost no difference in the selectivity observed experimentally from kcat and from kcat/KM. Thus we can also explore the difference in kcat (rather thankcat/KM). The contribution to the enantioselectivity in terms of kcat will be called here E':
| (3) |
where
| (4) |
In this work we will explore the selectivity by both strategies.
III. Computational Methods
The calculation of the activation free energies was performed by the empirical valence bond (EVB) method. This method that has been described extensively elsewhere20,21 is an empirical quantum mechanics/molecular mechanics (QM/MM) method46–49 that can be considered as a mixture of diabatic states describing the reactant(s), intermediate(s) and product(s) in a way that retains the correct change in structure and charge distribution along the reaction coordinate. The EVB diabatic states provide an effective way for evaluating the reaction free energy surface by using them for driving the system from the reactants to the product states in a free energy perturbation umbrella sampling procedure. The reason for the remarkable reliability of the EVB is, that it is calibrated on the reference solution reaction and then the calculations in the enzyme active site reflect (consistently) only the change of the environment21, exploiting the fact that the reacting system is the same in enzyme and solution. Thus, the EVB approach is calibrated only once in a study of a given type of enzymatic reaction.
The EVB for the present reaction has been constructed by using the three states described in Figure S1 (see supporting information). The EVB parameters for the surfaces of the solution reaction were calibrated by using the available experimental information about this reaction (see later in this section) and were kept unchanged for the generation of the protein EVB surface (see Tables S1 to S8 in the supporting information for detailed parameters, gas-phase shifts and off-diagonal elements).
The EVB calculations were carried out by the MOLARIS package50 using the ENZYMIX force field51. The EVB activation barriers were calculated at the configurations selected by the same free energy perturbation umbrella sampling (FEP/US) approach used in all our studies (e.g.21,52). The simulation systems were solvated by the surface constrained all atom solvent (SCAAS) model51 using a water sphere of 20 Å radius centered on the substrate and surrounded by a 2 Å grid of Langevin dipoles and then by a bulk solvent, while long-range electrostatic effects were treated by the local reaction field (LRF) method.51 The EVB region included the secondary alcohol substrate; the imidazole ring of HIS 224 and the entire side chain of Butanoyl-serine (see Figure S1 in the supporting information). Validation studies were done within 22 Å radius of inner sphere, where we repeated the calculations of the activation barrier and obtained practically the same results (treating the distanced ionized groups with a high dielectric macroscopic model). The FEP mapping procedure involved the use of 21 frames (5 ps each) for moving along the reaction coordinate and all the simulations were done at 300 K with a time step of 1fs.
In performing the calculations we found it useful to start with restraints and then to evaluate the free energy of releasing the restraints. The rational for this strategy is discussed and justified in section IV.4. The restraints used were aimed at keeping the oxygen of the alcohol in place, combined with a restraint that allows for the alcohol to reside within clear hydrogen bonding distance to the histidine base, as well as the asymmetric carbon atom of Butanoyl-serine.
While several sets of constraints were identified, all of which resulted in very similar free energy estimates, they also seemed to heavily affect the stability of the reaction throughout the simulation as small force constants were applied. The relation to the strong steric effects seen in CalB6,7,10,14,35,37,38 may well translate in seemingly “unconverged” or confined simulations, as more force is required to keep the reacting fragments in reasonable positions27. Thus, strong constraints of 10 kcal/mol were applied and the results presented here made use of a positional constraint for the oxygen of the secondary alcohol as well as two distance constraints keeping the oxygen within 3 A of the Nε of the HIS base as well as the carbonyl carbon of Butanoyl-serine. The simulations were repeated 4 times in order to obtain reliable results with different initial conditions (obtained from arbitrary points of the relaxation trajectory). Furthermore, the hysteresis in the calculation was examined by thermodynamic cycle closure (see section IV.3) and the average was determined by taking in each case the difference between the calculated minimum at the reaction state (RS) and the given transition state (TS). This simulation protocol was applied to reaction steps, the proton-transfer step and the nucleophilic attack.
The starting coordinates of the unbound CalB were obtained from the Protein Data Bank (PDB)53 (PDB ID 1TCA23), while the acylated serine was modeled based on the coordinates of a covalent phosphonate inhibitor (PDB ID 1LBS24). The same approach was used to model the mutant by simply mutating the tryptophan residue into an alanine using MOLARIS50. During system preparation, all crystal waters were removed while a structural water in clear hydrogen bonding distance to ASP187 was preserved (residue HOH 406 from the coordinates of PDB 1TCA23); all hydrogen atoms and all other water molecules were added using MOLARIS50,51 and the catalytic ASP187 was deprotonated in all simulations presented leading to a formal net charge of -1 for the protein (and not including the different states of the catalytically active HIS224).
The partial atomic charges for all resonance structures (see Figure S1 and Table S1 in the supporting information) were determined from the electronic wave functions by fitting the resulting electrostatic potential in the neighborhood of these molecules using the Merz-Singh-Kollman scheme.54 The electronic wave functions were calculated with hybrid density functional theory (DFT) using the B3LYP method55–58 and the 6–311G** basis set59–62, performed with the Gaussian03 package.63
The generated protein complex system (that includes the protein, bound ligand, water and Langevin dipoles) was pre-equilibrated for 1ns at 300K with a time steps of 1fs using the ENZYMIX force field.50,51 The spherical inner part of the system with radius 20Å was constrained by a weak harmonic potential of the form, with A=0.03 kcal mol−1Å2 to keep the protein atoms near the corresponding observed positions, along with the inner spherical constraints. The protein atoms outside this sphere were held fixed and their electrostatic effects excluded from the model.
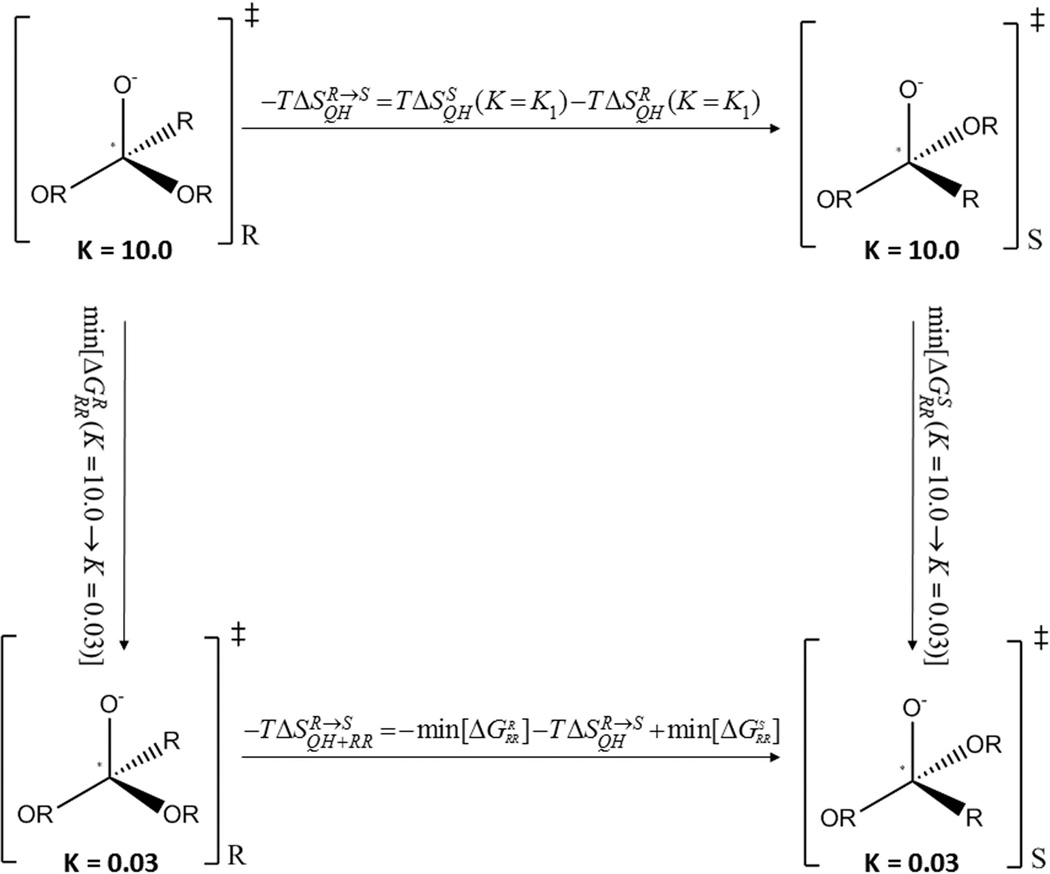
Since the present work includes a careful attempt to evaluate entropic contributions to enantioselectivity, we review here our restraint release (RR) approach for calculating entropies.64–66 (this approach should not be confused with the strategy of running the EVB with restraints in calculating the activation free energies). Our approach involves a variation of the original RR approach where we evaluate the entropy of a state with a potential UA, we start by adding to the potential of each state a strong harmonic constraint (with a force constant K set at 10.0 kcal−1 A−2) centered at a position, R̅, near the minimum of that potential. With a strong constraint we can evaluate the entropy of the system by the widely used quasi harmonic (QH) method67 (which is very problematic with realistic shallow potentials). Next we evaluate the entropy on the real potential by moving from the restraint potential to the real potential by releasing the strong restraint, K, and evaluating the corresponding free energy, ΔGRR. Here we use our finding that ΔGRR depends on the position of R̅, but the lowest ΔGRR obtained by changing R̅ includes only entropy and no enthalpic contributions (as was established in most details in64). Thus we can write
| (5) |
where ΔGRR designated the RR obtained by changing the indicated force constants and indicates the minimum value of the indicated ΔGRR from a set of different restraint coordinates (R̅). More specifically we apply a Cartesian restraint:
| (6) |
where i runs over the relevant coordinates (in the present case the solute coordinates ) and are reference coordinates that define the minimum of the restraining potential at the given state (e.g. N = I or N = II for the TS of the R and S-enantiomer respectively). Using this restraint we evaluate the QH entropy contribution, −T ΔS(K)OH for the energy of the system plus the restraint. We then evaluate the free energy of releasing the restraint using a standard FEP procedure with a mapping potential of the form
| (7) |
where E designates the unconstrained potential surface of the system and where λm is changed gradually from 0 to 1. Finally the relevant entropy is evaluated by Eq. 5. The calculations follow the simulation conditions of our EVB protocol with an 18 Å simulation sphere of explicit water molecules subject to the surface-constrained all-atom solvent boundary conditions. However, the RR-FEP involved the release of the position restraints between 48 windows, each with a simulation time of 40 ps at 300 K and a 1 fs time step.
IV. Results and Discussion
Our task is to quantify the energetics of the reaction for the R- and S-configured substrates in CalB where the modeled binding modes for the enantiomers are shown in Figure 4 (and will also be discussed in subsequent sections). With this initial structure we can calculate the relevant activation barriers as described below.
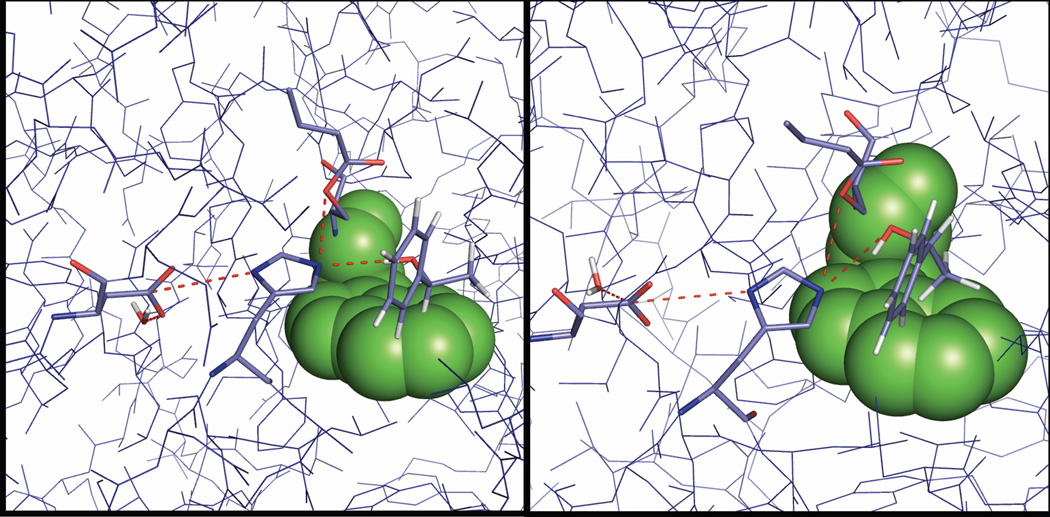
Figure 4.
A model of the RS shown after 1 ns equilibration for the R-enantiomer (left) and the S-enantiomer (right) (from the coordinates of PDB ID 1TCA23). The protein is shown in blue line representation with potential hydrogen bonds indicated by red dashed lines; the catalytic triad, together with phenylethanole and the preserved water molecule are shown in stick representation using light blue, navy and red for carbon, nitrogen and oxygen atoms, while TRP 104 acting as the proposed stereoselectivity pocket is shown in sphere representation using green. Hydrogens have been omitted for clarity and are only shown for the substrate as well as the preserved water molecule. The model was created using PyMol.81
IV. 1 Obtaining the EVB Surfaces for the Catalytic Reaction
Our study started with a systematic analysis of the reference reaction for histidine assisted ester hydrolysis in solution, using the relevant experimental information.21 The calibration procedure includes the energetics of the proton transfer (PT) from phenylethanole to imidazole and the energetics of the following nucleophilic attack (NA) of the ionized alcoholate and carbonyl carbon of butanoyl-serine (see ref7 and Figures 2 and 3).
As mentioned above, we start with the stepwise paths, separating the proton transfer and nucleophilic attack steps17,43. The energy of the proton transfer step (Figure 2) in water is determined from the pKa values (pKa (Phenylethanole) ~ 14.4 and pKa (His) ~721) and is found to be 10.1 kcal/mol.
The energy of forming the tetrahedral intermediate was calibrated by using the rate constant adapted from a similar uncatalyzed reaction. More specifically, we have considered the hydrolysis reaction of the hydroxide ion with methyl-isobutyrate and methyl-isopropionate - 18.6 and 18.9 kcal/mol respectively.68 While there is a common scaffold for these substrates to butanoyl-serine, our nucleophile is a secondary alcohol instead of the hydroxide ion. Here, an energetic difference of up to 4 kcal/mol of the activation barrier of alcoholate versus hydroxide was previously established on formamide esterification with specific hindsight on serine hydrolases.43 Combined with experimental and computational studies on similar substrates, all of which point towards an activation free energy between 15 and 19 kcal/mol32,68,69, we are confident in assuming a similar reduction for our specific reaction, which has subsequently been set at 15 kcal/mol for fitting the EVB solution surface.
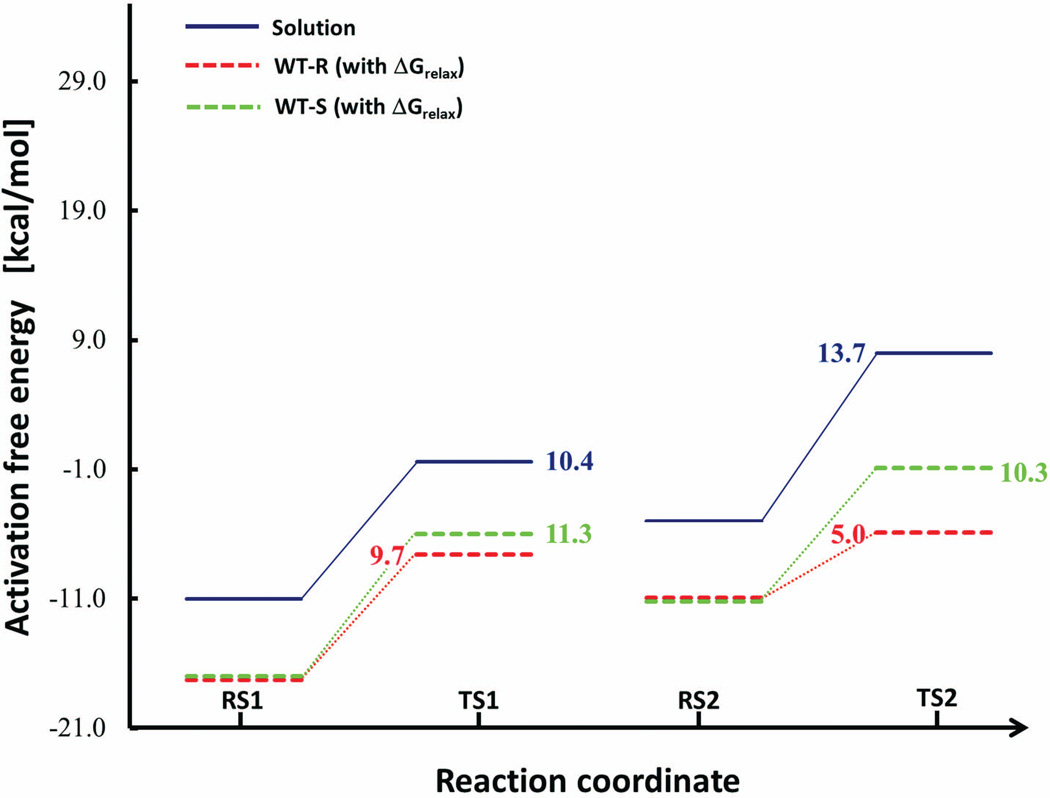
Combining the free energy for the PT step and the nucleophilic attack gives a total activation barrier () of ~25.1 kcal/mol for our reference reaction in the solvent cage. The barrier for a concerted path is expected to be very similar to our stepwise estimate (see ref17,43). The above estimate was used to calibrate the EVB surface for our reference reaction in solution and the corresponding free energy reference surfaces for the solution and the wild-type enzyme are shown in Figure 5. The results reported in the figure were obtained by a specialized restraint approach that will be discussed in section IV.4
Figure 5.
Diagram of the calculated activation barriers (in kcal/mol) for the transesterification using the EVB method. Here the final barriers are given that include in case of the enzyme the free energy of removing the restraints (not for the solution). The results reported in the figure were obtained by the specialized restraint relaxation approach that will be discussed in section IV.4
Using the EVB parameters calibrated on the reference solution reaction we evaluate the EVB free energy surface for the reaction in CalB. The resulting free energy calculations for each independent run are given in Tables II and III and IV for the results in solution, WT enzyme and mutant respectively. The final activation free energy estimates compared to experiment are summarized in Table V. As seen from Table V, we obtained a calculated activation barrier, Δgcat,calc, of 14.7 kcal/mol for the R-enantiomer and a barrier of 21.6 kcal/mol for the S-enantiomer. For completeness longer simulations were performed by extending the total runtime per frame three-fold and gave 15.3 kcal/mol for the R-enantiomer, hence lying within the statistical error of the method. Overall these barriers for the chemical reaction are in satisfying agreement with the observed barriers (Δgcat,exp=13.8 kcal/mol for the R-enantiomer and 22.1 for the S-enantiomer obtained using transition state theory respectively), and the observed kcat of the wild type CalB for the enantiomers.7
Table II.
Calculated activation free energies (in kcal/mol) for the reaction in solution (Sol) for four runs (b)
| Run | ΔGPT (Sol) | ΔGNA (Sol) | Δgcat,calc (Sol) |
| run1 | 10.1 | 13.5 | 23.6 |
| run2 | 11.7 | 14.6 | 26.3 |
| run3 | 9.8 | 12.9 | 22.7 |
| run4 | 10.3 | 13.6 | 23.9 |
| AVG | 10.4 | 13.7 | 24.1 |
| SD | 0.8 | 0.7 | 1.5 |
| MUE | 0.4 | 0.4 | 0.8 |
given by ΔGPT (Sol), ΔGNA (Sol) and Δgcat,calc (Sol) for the individual barriers of the PT and NA and the total calculated barrier respectively. Statistical figures of merit are given by the average (AVG), standard deviation (SD) and the mean unsigned error (MUE).
Table III.
Calculated activation free energies (in kcal/mol) for the reaction in the WT for four runs (c)
| Run | ΔGPT WT-R |
ΔGNA WT-R |
Δgcat,calc,relax WT-R |
ΔGPT WT-S |
ΔGNA WT-S |
Δgcat,calc,relax WT-S |
ΔΔgcat,calc,relax(a) WT (R→S) |
|---|---|---|---|---|---|---|---|
| 1 | 9.2 | 4.4 | 13.6 | 11.2 | 11.9 | 23.1 | 9.5 |
| 2 | 10.7 | 4.9 | 15.6 | 13.3 | 9.3 | 22.6 | 7.0 |
| 3 | 9.9 | 5.6 | 15.5 | 10.0 | 9.2 | 19.2 | 3.7 |
| 4 | 9.1 | 5.1 | 14.2 | 10.7 | 10.6 | 21.3 | 7.1 |
| AVE | 9.7 | 5.0 | 14.7 | 11.3 | 10.3 | 21.6 | 6.8 |
| SD | 0.4 | 0.7 | 1.0 | 1.4 | 1.0 | 2.5 | 2.4 |
| MUE | 0.2 | 0.3 | 0.5 | 0.7 | 0.5 | 1.2 | 1.2 |
The individual contributions ΔGPT and ΔGNA for each reaction and enantiomer provide the total barriers given by Δgcat,calc,relax. The absolute difference in activation free energies between the enantiomers was calculated with the free energy of releasing the geometrical restraints and is labelled ΔΔgcat,cal,relax. Statistical figures of merit are given by the average (AVG), standard deviation (SD) and the mean unsigned error (MUE).
Table IV.
Calculated activation free energies (in kcal/mol) for the reaction in the MUT for four runs (d)
| Run | ΔGPT MUT-R |
ΔGNA MUT-R |
Δgcat,calc MUT-R |
ΔGPT MUT-S |
ΔGNA MUT-S |
Δgcat,calc MUT-S |
ΔΔgcat,calc MUT (R→S) |
|---|---|---|---|---|---|---|---|
| 1 | 10.7 | 6.9 | 17.6 | 10.1 | 5.8 | 15.9 | −1.7 |
| 2 | 12.4 | 5.8 | 18.2 | 10.3 | 4.5 | 14.8 | −3.4 |
| 3 | 10.8 | 7.6 | 18.4 | 11.4 | 5.3 | 16.7 | −1.7 |
| 4 | 11.7 | 6.1 | 17.8 | 10.9 | 4.5 | 15.4 | −2.4 |
| AVE | 11.4 | 6.6 | 18.0 | 10.7 | 5.0 | 15.7 | −2.3 |
| SD | 0.8 | 0.8 | 0.4 | 0.6 | 1.1 | 0.8 | 0.8 |
| MUE | 0.4 | 0.4 | 0.2 | 0.3 | 0.6 | 0.4 | 0.4 |
The individual contribution ΔGPT and ΔGNA for each reaction and enantiomer provide the total barriers given by Δgcat,calc. The absolute difference in activation free energies between the enantiomers was calculated without the free energy of releasing the geometrical restraints and is labeled ΔΔgcat,calc. Statistical figures of merit are given by the average (AVG), standard deviation (SD) and the mean unsigned error (MUE).
Table V.
Calculated activation free energies (in kcal/mol) for the PT and NA reactions in the WT and the MUT for each enantiomer.(e)
| System | ΔGPT | ΔGNA | Δgcat,calc(a) | Δgcat,calc,relax(b) | Δgcat,exp(c) |
|---|---|---|---|---|---|
| solution R/S | 10.4 | 13.7 | 24.1 | na | 25.1 |
| WT-R | 9.7 | 5.0 | 14.7 | 16.7 | 13.8 |
| WT-S | 11.3 | 10.3 | 21.6 | 23.9 | 22.1 |
| MUT-R | 11.4 | 6.6 | 18.0 | na | 16.7 |
| MUT-S | 10.7 | 5.0 | 15.7 | na | 15.5 |
The total barriers without and with addition of the free energy of removing the geometrical restraints at the RS and TS are indicated by Δgcat,cal and Δgcat,calc,Relax respectively, and the experimental barrier is given by Δgcat,exp. Statistical figures of merit are given by the average (AVG), standard deviation (SD) and the mean unsigned error (MUE).
IV. 2 The Binding of Secondary Alcohols to Candida Antarctica lipase B
An issue that has played an important role in the discussion of the selectivity of CalB and other enzymes is the nature of the binding modes6,7,10,14,24,27,28,31,35,38. Although this issue appears to be overemphasized due to the use of approaches with limited sampling (see below), it is very useful to consider this issue here.
The empirical Kazlauskas rule76 has been used to assess the most likely binding mode of the fast-reacting enantiomer. That is, while the rule suggests a clear preference for the (R)-enantiomer of a secondary alcohol, which seems confirmed by experiment8,17,32, it makes modeling the (S)-enantiomer almost impossible, as its’ large phenyl ring cannot fit in the postulated stereoselectivity pocket, providing no significant conformational changes are involved, while for the R-enantiomer the large group would point towards bulk solvent. This is illustrated in Figure 6 where the binding mode on the right is termed "unproductive" as the medium-sized substituent binds in the larger stereoselectivity pocket, while the "productive" mode, shown on the left, would be able to accommodate the larger substituent and facilitate catalysis.
Figure 6.
The productive and unproductive binding modes proposed for the binding of secondary alcohols to CalB. While the binding mode shown on the left can easily accommodate both substituents, the binding mode on the right cannot accommodate the large substituent L in the postulated pocket, thus making it the slow-reacting enantiomer according to Kazlauzkas.76
The asymmetric sp3-hybridized carbon atom binds four substituents, three of which define its absolute configuration (relative to the hydrogen in our case), which amounts, theoretically, to a total of 4 different binding modes for the S-enantiomer. According to Cahn-Ingold-Prelog70,71, these were systematically assigned according to the weight of their substituents, thus leading to H/O, M/L and M/H permutations compared to the R-enantiomer (Here H, O, M and L refer to hydrogen, oxygen, medium-sized and large-sized substituents, see for example ref28).
The interconversion between those modes during a simulation is subject to the choice of sampling, which in turn is combined with the natural confinement presented by the detailed architecture of the active site. Here, the use of restraints to maintain a particular binding mode during the simulation – or simply just a certain geometry - is a useful tool. However, as it will affect the free energies by modifying the conformational sampling, it becomes necessary to evaluate the free energy of removing the restraint(s). The effectiveness of this approach will be illustrated in section IV.4.
IV.3. Calculating the Selectivity In Terms of TS Binding energy
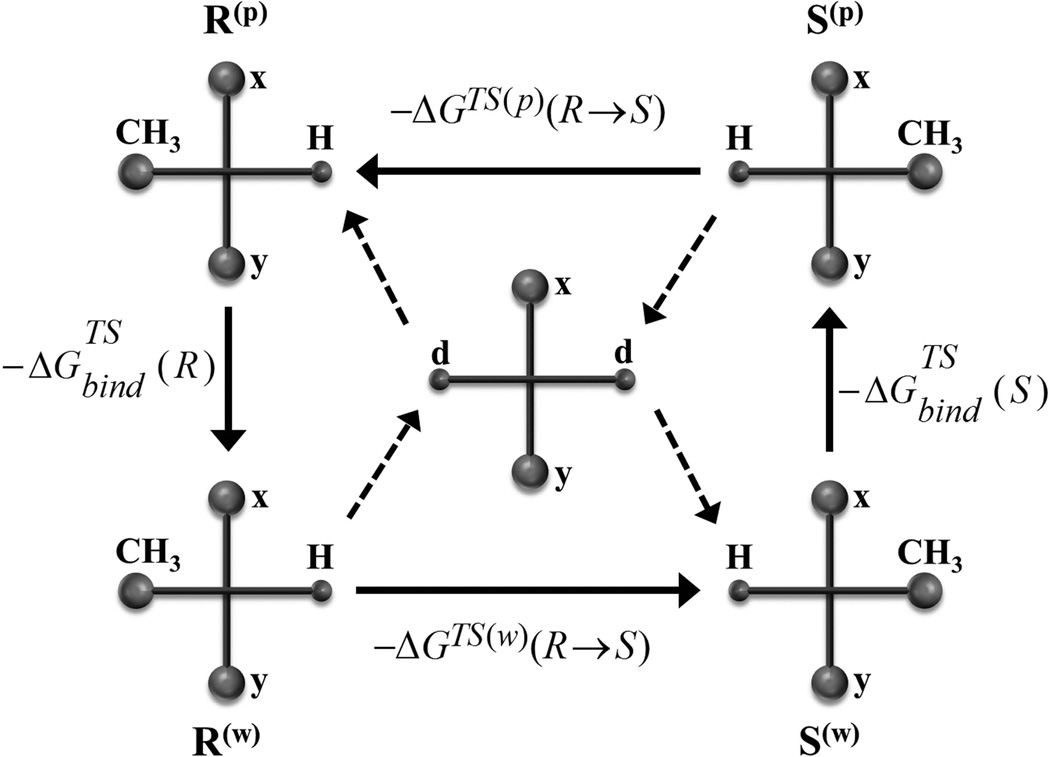
The rate-limiting step in the transesterification studied here is the second nucleophilic attack of the secondary alcoholate on the carbonyl carbon of the modified serine residue8,17,32. Consequently, the enantioselectivity can be defined as the difference between the TS binding free energies of the R- and S enantiomer. As outlined previously22, we can use the thermodynamic cycle of Figure 7 and obtain:
| (8) |
Figure 7.
Thermodynamic cycle for the mutation of the R-enantiomer into the S-enantiomer. Here, the mutation of CH3 to H and H to CH3 is described schematically. As outlined in the main text, the mutation is practically done at the transition state by converting both, the CH3 and H, to dummy atoms.
In using this expression it is enough to mutate the R to S in the TS in the protein and then just substract the corresponding results for the mutation in water (this reflects force field artifacts). The approach of mutating the TS has already been used in our early mutational studies72, but at that time the mutation was performed on the protein, while here we mutate the substrate. This is achieved by converting the groups that distinguish R from S to real or dummy atoms. More specifically, we mutate the dummy atoms in R to real atoms in S and the real atoms in R to dummy atoms in S.
The reaction coordinate for moving from the real TS to the dummy TS was split into 31 frames (of 5ps each) for the FEP mapping procedure and all the simulations were performed at 300K with a 1 fs time step. The structures used were taken from the corresponding EVB calculations, after a further relaxation for 800ps for each system, and each simulation was repeated 8 times in order to obtain reliable results with different initial conditions (obtained from arbitrary points along the TS relaxation trajectory). In order to avoid the end-point catastrophe in mutating dummy atoms, we found it convenient to delete the first and last frame, while exploring the convergence behavior with larger number of frames.
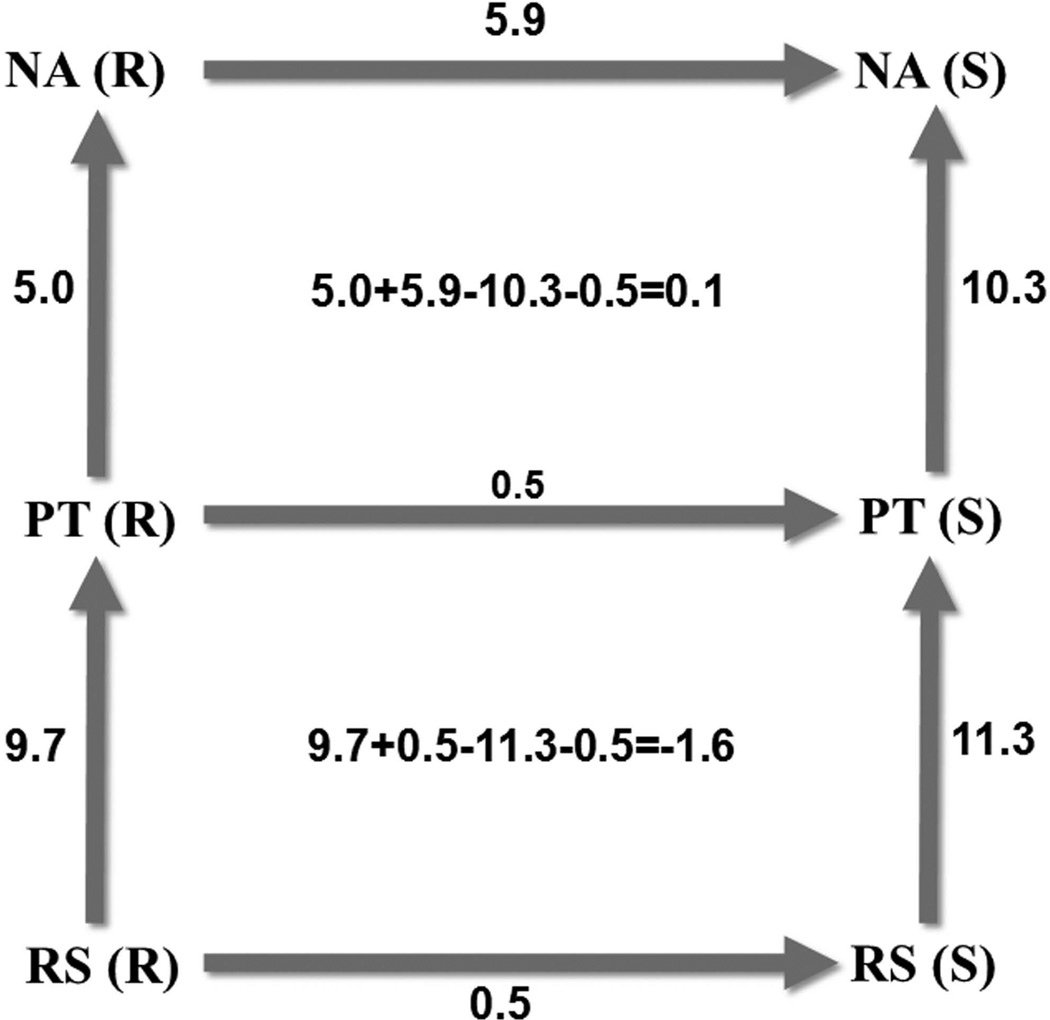
The results of these R to S mutations at the TS are summarized in Table VI. While the agreement with experiment is reasonable (the absolute error lies around 2 kcal/mol for the wild-type CalB), the results are interesting since they allow us to complete thermodynamic cycles that involve not only the rate-limiting step, but the entire chemical reaction. That is, we perform the same procedure for the PT step to calculate the binding free energy of the TS for the proton transfer, and finally calculate the energy for perturbing the atoms distinguishing R from S at the RS using our standard EVB procedure (also by perturbing the atoms distinguishing R from S to dummy atoms). While no experimental information is available for these (the experimental observation is based on the rate-limiting step) they still allow us to construct probably the first complete thermodynamic cycle over an entire chemical reaction. Overall the energies given along the cycle in Figure 8 provide a major consistency check on our approach.
Table VI.
Calculated binding free energies (in kcal/mol) of the TS(f)
| System | Selectivity | ΔΔGbind,calc(TS) | ΔΔGbind,exp(TS) |
|---|---|---|---|
| WT/PT | R | 0.5±1.6 | 8.3 |
| WT/NA | R | 5.9±2.0 | |
| MUT/PT | R | 0.5±1.3 | −1.4 |
| MUT/NA | R | 3.5±2.2 |
ΔΔGbind,calc (TS) is compared to experiment ΔΔGbind,exp (TS) for the PT and the NA bound to the WT and the MUT. The preferred enantiomer is given in the column denoted by “Selectivity;” the calculated ΔΔGbind,calc (TS) is given with the MUE over a total of 8 independent runs.
Figure 8.
Thermodynamic cycle over the entire chemical reaction. For the vertical arrows the EVB method is used to estimate the activation free energies responsible for catalysis, while the horizontal arrows stand for the mutations (to dummy atoms) aiming to calculate the binding free energies of the TS of each of the chemical reactions, i.e. for the PT as well as the NA for each enantiomer (in brackets). To be able to close the lower cycle, mutations have also been performed on the RS, but here the EVB method has been used to turn the respective atoms into dummies. The numbers in the center of each cycle indicate the observed hysteresis effects, standing at 0.1 for the NA and −1.6 for the PT. All units are in kcal/mol.
IV. 4. Obtaining Improved Result by Introducing and Removing Restraints
The involvement of large steric effects in the enantioselective recognition by CalB was found to present a significant challenge to calculations of the relevant free energies. That is, we found that running simulations form different initial conditions tends to give rather unstable results for the calculated enantioselectivity. On the other hand, performing free energy calculations with restraint and then releasing the restraint may eliminate the need for extensively long simulations. That is, calculations with restraints tend to be stable since they are confined to a well-defined region of the configurational space and the relaxation from the restraint region tends to give more stable results than those obtained by staring form arbitrary configurations in a shallow landscape.
A comparison of the calculated free energies obtained for the PT and NA run at K set to 0.03 kcal/mol would at least support the above claim. Clearly, inspection of Tables III and IV shows an expected mean unsigned error (MUE) for the final barriers well within 1.2 kcal/mol which rapidly degrades to 2.5 for the final barriers obtained with simulations run at K=0.03 kcal/mol (see Table VII). Consequently, the free energy of removing the restraint(s) allows us to estimate the “relaxation” free energies, ΔGrelax, and “relax” the final barriers accordingly. Therefore we start with the free energies of the RS and the TS of the PT and the NA obtained by our EVB simulations that were run at K set to 10.0 kcal mol−1 A−2 and then add the correction obtained upon reducing K to 0.03 over a total of 48 evenly spaced Ks. The same value was also used for K1 in our entropy calculations. For consistency, all other MD simulation conditions detailed in section III are maintained, as were the free energies calculated using the standard FEP procedure. The underlying thermodynamic cycle for calculating Δgcat,calc,relax shown in Figure S2 (supporting information) illustrates our approach and the obtained barriers for the PT and NA are given in Figure 6 for the WT (or a more detailed presentation in Figure S3 in the supporting information). Interestingly the average values of the results did not change drastically (see Table VII) but the stability of the calculations deteriorated very significantly.
Table VII.
Calculated activation free energies (in kcal/mol) for the reaction in the WT for four runs (g)
| Run | ΔGPT WT-R | ΔGNA WT-R | ΔGPT WT-S | ΔGNA WT-S | ΔΔgcat,cal (R→S) |
|---|---|---|---|---|---|
| 1 | 12.4 | 5.6 | 10.2 | 14.5 | 6.7 |
| 2 | 9.1 | 14.4 | 12.5 | 8.5 | −2.5 |
| 3 | 14.1 | 3.5 | 15.4 | 6.5 | 4.3 |
| 4 | 9.5 | 4.5 | 5.8 | 17.5 | 9.3 |
| AVG | 11.3 | 7.0 | 10.9 | 11.7 | 4.5 |
| SD | 2.4 | 5.0 | 4.1 | 5.1 | 5.1 |
| MUE | 1.2 | 2.5 | 2.0 | 2.6 | 2.5 |
using K=0.03 for the geometrical constraints. The difference in individual contribution ΔGPT and ΔGNA for each reaction and enantiomer gives the absolute difference in activation free energies between the enantiomers, ΔΔgcat,calc (R→S). Statistical figures of merit are given by the average (AVG), standard deviation (SD) and the mean unsigned error (MUE).
IV. 5 Exploring Entropic Contributions to Selectivity
Following the outline in the method section we conducted calculations of the entropic difference between the enantiomers at the TS. This was done using the two paths according to the cycle shown in Figure 9 and our calculated results given in Table VIII for both enantiomers bound to WT CalB.
Figure 9.
Thermodynamic cycle used to estimate the entropic contribution to catalysis for binding the TS of the enantiomers. The vertical arrows correspond to the simulations where the RR approach is used to perturb/release the restraint, the QH approximation is utilized to calculate the contribution from infinite to strong restraint (top horizontal arrow).
Table VIII.
Calculated entropies (in kcal/mol) provided by the RR and QH calculations for the NA at the TS (h)
| WT-R (TS) | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| RR 10 →0.03 | −35.3 | −35.8 | −35.9 | −36.2 |
| QH10 | −29.8 | −29.5 | −30.0 | −29.5 |
| RR+QH | −65.1 | −65.3 | −65.9 | −65.9 |
| WT-S (TS) | 1 | 2 | 3 | 4 |
| RR10 →0.03 | −35.5 | −34.9 | −35.2 | −35.1 |
| QH10 | −28.7 | −28.9 | −29.4 | −28.5 |
| RR+QH | −64.2 | −63.8 | −64.6 | −63.6 |
| MUT-S (TS) | 1 | 2 | 3 | 4 |
| RR10 →0.03 | −32.2 | −33.5 | −32.9 | −32.6 |
| QH10 | −27.4 | −27.9 | −27.6 | −27.3 |
| RR+QH | −59.6 | −61.4 | −60.5 | −59.9 |
| MUT-R (TS) | 1 | 2 | 3 | 4 |
| RR10 →0.03 | −33.5 | −33.9 | −34.0 | −33.3 |
| QH10 | −30.8 | −30.5 | −31.0 | −30.8 |
| RR+QH | −64.3 | −64.4 | −65.0 | −63.8 |
The table includes the results over four (1 to 4) simulations, each using different restraint coordinates. Each simulation consisted of 48 windows, over 40ps at 300K and a 1fs time step. The bold values indicate the calculated minimum, their difference reflecting the best estimate of -TΔS. As discussed in the main text, this variational minimization reflects the fact that all the RR free energies contain enthalpic contributions, which approach zero for restraint coordinates that gives the lowest RR/QH energies.
Our practical RR calculations involved the following steps: after an initial relaxation using MD runs of 1ns at 300 K with a time step of 1fs, arbitrary points along the trajectory were then collected to generate eight different sets of Rs, the constraint coordinates. The RR contributions for each of these sets were then evaluated. This was done by calculating the QH contribution with =10.0 kcal mol−1A−2 using
| (9) |
Where ΔSB (K = K1)QH designates the term of Eq. 5 for the potential B by in the presence of the constraint.
The calculated results are given in Table VIII for both enantiomers bound to the W104A mutant. The reported results were obtained by releasing the constraints on the TS of the substrates atoms while leaving the enzyme unconstraint. A more rigorous treatment should involve also a RR treatment of the active site65,73, but it is likely that in the present case exploring the entropy of the substrate with a relaxed enzyme is a reasonably reliable option. Encouragingly, we obtained 3.5 kcal/mol difference in the entropic contribution whereas the experiment in CH3CN, for example, lies at 5.0 (or 8.1 and 8.4 kcal/mol in decaline and cyclohexane respectively). Here, the obvious difference lies in the fact that organic media are used in the experiment, while the simulation is performed in water. Arguably, this difference in the entropic contribution may well reflect the subtle protein conformational change induced by the solvent (and temperature), which finally translates energetically to catalysis. In this case, our results would suggest that the W104A mutant of CalB is likely to share the most important conformational features between water and CH3CN, and that the imposition of constraints on all atoms of the evb region is sufficient for estimating the entropic contribution to catalysis. Nevertheless, to quantify accurately the impact of protein flexibility and conformational states in different solvents (and with respect to the entropy at different temperatures) to catalysis, more detailed studies are required in combination with parameterization of novel solvent models for biomolecular simulations.
Finally, and as shown previously64,73, a more rigorous treatment of the entropy ideally involves the RR on the entire active site. Here one may assume that in the particular case of CalB the entropic difference, that is the difference between the R and S enantiomer at the TS, is sufficient to provide us with an accurate estimate for the entropic contribution, and that the extension of the RR to the entire active site will not change the absolute difference between the enantiomers at the TS dramatically. However, this was not attempted in this work, as the main focus here lies on calculating the enantioselectivity on a real biological system, while the encouraging results of the entropy calculations certainly provide us with a benchmark suggesting to focus our RR approach more systems.
V. Concluding Remarks
Optimizing enzymes to catalyze selective enantioselective reactions has a major potential in biotechnology1,74–78, including in the generation of biocatalysts for efficient synthesis of enantiomerically pure chiral molecules for the production of drugs by the pharmaceutical industry74,79,80.
Recent advances in this field have been due in part to directed evolution experiments11,74–77,79,80 with some qualitative insight from theoretical studies (see Introduction). It seems to us that at the present stage it is important to push the capacity of theoretical simulation as useful tool in designing enantioselectivity enzymes. While previous studies9,13,16,17,27,28,31,32,36,69, including ours22, gave encouraging results, the ability of simulations to model consistently and reliably strain effects on enantioselectivity have not been demonstrated. In fact, a recent study27 of the WT CalB was forced to use a very large constraint of 100 kcal mol−1 A−2 to guarantee a hydrogen bond between the substrate and the imidazole base. Of course, using such a treatment cannot be a justifiable strategy. Similarly, energy minimization strategies are unlikely to allow proper relaxation of the protein steric forces. This does not mean that simple energy minimization, and even just visual inspection, cannot give a powerful guide for getting effective mutations. However, approaches that do not involve extensive sampling are unlikely to give stable results, just because of the fact that starting from different initial configurations gives different results. In principle one should be able to use free energy calculations, but the reliability of such strategies must be established and this has been the main focus of the present work.
The present study confirmed the suspicion that a converging prediction of enantioselectivity is a major challenge (since major sampling is needed to obtain converging results). Our results demonstrate reasonable thermodynamic cycle closure, providing us with yet more confidence and validation of our method and established formally rigorous free energy calculation of mutational effects of absolute enantioselectivity, where all the elements of the calculation involve careful sampling of what is basically a reliable semiempirical QM/MM potential over the entire chemical reaction.
In addition to the demonstration that we can capture reliably steric contributions to enantioselectivity, we also explored here our ability to evaluate the corresponding entropic effects. It appears that the RR approach coupled with the EVB offers a powerful way of accomplishing this task. Further studies should focus on the change in entropy with the change of the solvent as this remarkable effect may give interesting clues on the role of organic solvents in catalysis.
In view of the effectiveness of the present approach we hope that it can serve as a general tool for screening and refining enantioselective enzymes.
Supplementary Material
Acknowledgments
We like to thank Maria P. Frushicheva for useful discussions and valuable comments. This work was supported by Grant GM024492 from the National Institutes of Health (NIH). We thank the University of Southern California´s High Performance Computing and Communication Center (HPCC) for computer time.
References
- 1.Beck G. Synthesis of chiral drug substances. Synlett. 2002;6:837–850. [Google Scholar]
- 2.Breuer M, Ditrich K, Habicher T, Hauer B, Keßeler M, Stürmer R, Zelinski T. Industrial methods for the production of optically active intermediates. Angewandte Chemie International Edition. 2004;43(7):788–824. doi: 10.1002/anie.200300599. [DOI] [PubMed] [Google Scholar]
- 3.Engström K, Nyhlén J, Sandström AG, Bäckvall J-E. Directed evolution of an enantioselective lipase with broad substrate scope for hydrolysis of α-substituted esters. Journal of the American Chemical Society. 2010;132(20):7038–7042. doi: 10.1021/ja100593j. [DOI] [PubMed] [Google Scholar]
- 4.Prasad S, Bocola M, Reetz MT. Revisiting the lipase from Pseudomonas aeruginosa: Directed evolution of substrate acceptance and enantioselectivity using iterative saturation mutagenesis. ChemPhysChem. 2011;12(8):1550–1557. doi: 10.1002/cphc.201100031. [DOI] [PubMed] [Google Scholar]
- 5.Reetz MT, Prasad S, Carballeira JD, Gumulya Y, Bocola M. Iterative saturation mutagenesis accelerates laboratory evolution of enzyme stereoselectivity: Rigorous comparison with traditional methods. Journal of the American Chemical Society. 2010;132(26):9144–9152. doi: 10.1021/ja1030479. [DOI] [PubMed] [Google Scholar]
- 6.Magnusson AO, Rotticci-Mulder JC, Santagostino A, Hult K. Creating space for large secondary alcohols by rational redesign of Candida antarctica lipase B. ChemBioChem. 2005;6(6):1051–1056. doi: 10.1002/cbic.200400410. [DOI] [PubMed] [Google Scholar]
- 7.Magnusson AO, Takwa M, Hamberg A, Hult K. An S-selective lipase was created by rational redesign and the enantioselectivity increased with temperature. Angewandte Chemie International Edition. 2005;44(29):4582–4585. doi: 10.1002/anie.200500971. [DOI] [PubMed] [Google Scholar]
- 8.Martinelle M, Hult K. Kinetics of acyl transfer reactions in organic media catalysed by Candida antarctica lipase B. Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology. 1995;1251(2):191–197. doi: 10.1016/0167-4838(95)00096-d. [DOI] [PubMed] [Google Scholar]
- 9.Ottosson J, Fransson L, Hult K. Substrate entropy in enzyme enantioselectivity: An experimental and molecular modeling study of a lipase. Protein Science. 2002;11(6):1462–1471. doi: 10.1110/ps.3480102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rotticci D, Hæffner F, Orrenius C, Norin T, Hult K. Molecular recognition of sec-alcohol enantiomers by Candida antarctica lipase B. Journal of Molecular Catalysis B: Enzymatic. 1998;5(1–4):267–272. [Google Scholar]
- 11.Sandström AG, Engström K, Nyhlén J, Kasrayan A, Bäckvall J-E. Directed evolution of Candida antarctica lipase A using an episomaly replicating yeast plasmid. Protein Engineering Design and Selection. 2009;22(7):413–420. doi: 10.1093/protein/gzp019. [DOI] [PubMed] [Google Scholar]
- 12.Sandström AG, Wikmark Y, Engström K, Nyhlén J, Bäckvall J-E. Combinatorial reshaping of the Candida antarctica lipase A substrate pocket for enantioselectivity using an extremely condensed library. Proceedings of the National Academy of Sciences. 2012;109(1):78–83. doi: 10.1073/pnas.1111537108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nyhlén J, Martín-Matute B, Sandström AG, Bocola M, Bäckvall J-E. Influence of δ-functional groups on the enantiorecognition of secondary alcohols by Candida antarctica lipase B. ChemBioChem. 2008;9(12):1968–1974. doi: 10.1002/cbic.200800036. [DOI] [PubMed] [Google Scholar]
- 14.Orrenius C, Haeffner F, Rotticci D, Ohrner N, Norin T, Hult K. Chiral recognition of alcohol enantiomers in acyl transfer reactions catalysed by Candida antarctica lipase B. Biocatalysis and Biotransformation. 1998;16(1):1–15. [Google Scholar]
- 15.Léonard V, Fransson L, Lamare S, Hult K, Graber M. A water molecule in the stereospecificity pocket of Candida antarctica lipase B enhances enantioselectivity towards pentan-2-ol. ChemBioChem. 2007;8(6):662–667. doi: 10.1002/cbic.200600479. [DOI] [PubMed] [Google Scholar]
- 16.Raza S, Fransson L, Hult K. Enantioselectivity in Candida antarctica lipase B: A molecular dynamics study. Protein Science. 2001;10(2):329–338. doi: 10.1110/ps.33901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Svedendahl M, Carlqvist P, Branneby C, Allnér O, Frise A, Hult K, Berglund P, Brinck T. Direct epoxidation in Candida antarctica lipase B studied by experiment and theory. ChemBioChem. 2008;9(15):2443–2451. doi: 10.1002/cbic.200800318. [DOI] [PubMed] [Google Scholar]
- 18.Hopmann KH, Hallberg BM, Himo F. Catalytic mechanism of limonene epoxide hydrolase: A theoretical study. Journal of the American Chemical Society. 2005;127(41):14339–14347. doi: 10.1021/ja050940p. [DOI] [PubMed] [Google Scholar]
- 19.Siegbahn PEM, Himo F. The quantum chemical cluster approach for modeling enzyme reactions. Wiley Interdisciplinary Reviews: Computational Molecular Science. 2011;1(3):323–336. [Google Scholar]
- 20.Aqvist J, Warshel A. Simulation of enzyme-reactions using valence-bond force-fields and other hybrid quantum-classical approaches. Chemical Reviews. 1993;93(7):2523–2544. [Google Scholar]
- 21.Warshel A. Computer modeling of chemical reactions in enzymes and solutions.: Wiley-Interscience. 1991 [Google Scholar]
- 22.Frushicheva MP, Warshel A. Towards quantitative computer-aided studies of enzymatic enantioselectivity: The case of candida antarctica lipase A. ChemBioChem. 2012;13(2):215–223. doi: 10.1002/cbic.201100600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uppenberg J, Hansen M, Patkar S, Jones T. The sequence, crystal structure determination and refinement of two crystal forms of lipase B from Candida antarctica. Structure. 1994;2(4):293–308. doi: 10.1016/s0969-2126(00)00031-9. [DOI] [PubMed] [Google Scholar]
- 24.Uppenberg J, Oehrner N, Norin M, Hult K, Kleywegt GJ, Patkar S, Waagen V, Anthonsen T, Jones TA. Crystallographic and molecular-modeling studies of lipase B from Candida antarctica reveal a stereospecificity pocket for secondary alcohols. Biochemistry. 1995;34(51):16838–16851. doi: 10.1021/bi00051a035. [DOI] [PubMed] [Google Scholar]
- 25.Egmond MR, van Bemmel CJ. Impact of structural information on understanding lipolytic function. In: Byron Rubin EAD, editor. Methods in Enzymology. Volume 284. Academic Press; 1997. pp. 119–129. Volume. [DOI] [PubMed] [Google Scholar]
- 26.Schrag JD, Cygler M. Lipases and α-β hydrolase fold. In: Byron Rubin EAD, editor. Methods in Enzymology. Volume Volume 284. Academic Press; 1997. pp. 85–107. [DOI] [PubMed] [Google Scholar]
- 27.Chaput L, Sanejouand YH, Balloumi A, Tran V, Graber M. Contribution of both catalytic constant and Michaelis constant to CALB enantioselectivity: Use of FEP calculations for prediction studies. Journal of Molecular Catalysis B: Enzymatic. 2012;76(0):29–36. [Google Scholar]
- 28.Xu T, Zhang L, Su E, Cui D, Wang X, Wei D. Disparity in productive binding mode of the slow-reacting enantiomer determines the novel catalytic behavior of Candida antarctica lipase B. Journal of Molecular Catalysis B: Enzymatic. 2010;62(3–4):288–296. [Google Scholar]
- 29.Bocola M, Otte N, Jaeger K-E, Reetz MT, Thiel W. Learning from directed evolution: Theoretical investigations into cooperative mutations in lipase enantioselectivity. ChemBioChem. 2004;5(2):214–223. doi: 10.1002/cbic.200300731. [DOI] [PubMed] [Google Scholar]
- 30.de los Ríos AP, Hernández-Fernández FJ, Tomás-Alonso F, Gómez D, Víllora G. Biocatalytic kinetic resolution of rac-1-phenylethanol and rac-2-pentanol in hexane medium: Acyl donor and water content effects. The Canadian Journal of Chemical Engineering. 2010;88(3):442–446. [Google Scholar]
- 31.Hæffner F, Norin T, Hult K. Molecular modeling of the enantioselectivity in lipase-catalyzed transesterification reactions. Biophysical Journal. 1998;74(3):1251–1262. doi: 10.1016/S0006-3495(98)77839-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwon HC, Shin DY, Lee JH, Kim SW, Kang JW. Molecular modeling and its experimental verification for the catalytic mechanism of Candida antarctica lipase B. 2007:1098–1105. [PubMed] [Google Scholar]
- 33.Léonard-Nevers V, Marton Z, Lamare S, Hult K, Graber M. Understanding water effect on Candida antarctica lipase B activity and enantioselectivity towards secondary alcohols. Journal of Molecular Catalysis B: Enzymatic. 2009;59(1–3):90–95. [Google Scholar]
- 34.Li C, Tan T, Zhang H, Feng W. Analysis of the conformational stability and activity of candida antarctica lipase B in organic solvents insight from molecular dynamics and quantum mechanics/molecular mechanics simulations. Journal of Biological Chemistry. 2010 doi: 10.1074/jbc.M110.136200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marton Z, Léonard-Nevers V, Syrén PO, Bauer C, Lamare S, Hult K, Tranc V, Graber M. Mutations in the stereospecificity pocket and at the entrance of the active site of Candida antarctica lipase B enhancing enzyme enantioselectivity. Journal of Molecular Catalysis B: Enzymatic. 2010 [Google Scholar]
- 36.Ottosson J, Fransson L, King JW, Hult K. Size as a parameter for solvent effects on Candida antarctica lipase B enantioselectivity. Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology. 2002;1594(2):325–334. doi: 10.1016/s0167-4838(01)00324-7. [DOI] [PubMed] [Google Scholar]
- 37.Parida S, Dordick JS. Tailoring lipase specificity by solvent and substrate chemistries. The Journal of Organic Chemistry. 1993;58(12):3238–3244. [Google Scholar]
- 38.Vallin M, Syrén P-O, Hult K. Mutant lipase-catalyzed kinetic resolution of bulky phenyl alkyl sec-alcohols: A thermodynamic analysis of enantioselectivity. ChemBioChem. 2010;11(3):411–416. doi: 10.1002/cbic.200900635. [DOI] [PubMed] [Google Scholar]
- 39.Wittrup Larsen M, Zielinska DF, Martinelle M, Hidalgo A, Jensen LJ, Bornscheuer UT, Hult K. Suppression of water as a nucleophile in Candida antarctica lipase B catalysis. ChemBioChem. 2010;11(6):796–801. doi: 10.1002/cbic.200900743. [DOI] [PubMed] [Google Scholar]
- 40.Warshel A, Russell S. Theoretical correlation of structure and energetics in the catalytic reaction of trypsin. Journal of the American Chemical Society. 1986;108(21):6569–6579. [Google Scholar]
- 41.Warshel A, Narayszabo G, Sussman F, Hwang JK. How do serine proteases really work. Biochemistry. 1989;28(9):3629–3637. doi: 10.1021/bi00435a001. [DOI] [PubMed] [Google Scholar]
- 42.Creighton S, Hwang JK, Warshel A, Parson WW, Norris J. Simulating the dynamics of the primary charge separation process in bacterial photosynthesis. Biochemistry. 1988;27(2):774–781. [Google Scholar]
- 43.Strajbl M, Florián J, Warshel A. Ab initio evaluation of the potential surface for general base-catalyzed methanolysis of formamide: A reference solution reaction for studies of serine proteases. Journal of the American Chemical Society. 2000;122(22):5354–5366. [Google Scholar]
- 44.Ishikita H, Warshel A. Predicting drug-resistant mutations of HIV protease. Angewandte Chemie International Edition. 2008;47(4):697–700. doi: 10.1002/anie.200704178. [DOI] [PubMed] [Google Scholar]
- 45.Warshel A, Sussman F, King G. Free energy of charges in solvated proteins: Microscopic calculations using a reversible charging process. Biochemistry. 1986;25(26):8368–8372. doi: 10.1021/bi00374a006. [DOI] [PubMed] [Google Scholar]
- 46.Strajbl M, Hong G, Warshel A. Ab initio QM/MM simulation with proper sampling: “First Principle” calculations of the free energy of the autodissociation of water in aqueous solution. The Journal of Physical Chemistry B. 2002;106(51):13333–13343. [Google Scholar]
- 47.Shurki A, Warshel A. Structure/function correlations of proteins using MM, QM/MM, and related approaches: Methods, concepts, pitfalls, and current progress. PROTEIN SIMULATIONS. 2003;66:249–313. doi: 10.1016/s0065-3233(03)66007-9. [DOI] [PubMed] [Google Scholar]
- 48.Ranaghan KE, Mulholland AJ. Investigations of enzyme-catalysed reactions with combined quantum mechanics/molecular mechanics (QM/MM) methods. International Reviews in Physical Chemistry. 2010;29(1):65–133. [Google Scholar]
- 49.Senn HM, Thiel W. QM/MM Methods for Biomolecular Systems. Angewandte Chemie International Edition. 2009;48(7):1198–1229. doi: 10.1002/anie.200802019. [DOI] [PubMed] [Google Scholar]
- 50.Warshel A. MOLARIS-XG: Theoretical background and practical examples. 2013 available at http://laetro.usc.edu/programs/doc/theory_molaris_911pdf. [Google Scholar]
- 51.Lee FS, Chu ZT, Warshel A. Microscopic and semimicroscopic calculations of electrostatic energies in proteins by the Polaris and Enzymix programs. Journal of Computational Chemistry. 1993;14(2):161–185. [Google Scholar]
- 52.Warshel A, Sharma PK, Kato M, Xiang Y, Liu H, Olsson MHM. Electrostatic basis for enzyme catalysis. Chemical Reviews. 2006;106(8):3210–3235. doi: 10.1021/cr0503106. [DOI] [PubMed] [Google Scholar]
- 53.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Research. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh UC, Kollman PA. An approach to computing electrostatic charges for molecules. Journal of Computational Chemistry. 1984;5(2):129–145. [Google Scholar]
- 55.Becke AD. Density-functional thermochemistry. III. The role of exact exchange. The Journal of Chemical Physics. 1993;98(7):5648–5652. [Google Scholar]
- 56.Lee C, Yang W, Parr RG. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Physical Review B. 1988;37(2):785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 57.Stephens PJ, Devlin FJ, Chabalowski CF, Frisch MJ. Ab initio calculation of vibrational absorption and circular dichroism spectra using density functional force fields. The Journal of Physical Chemistry. 1994;98(45):11623–11627. [Google Scholar]
- 58.Vosko SH, Wilk L, Nusair M. Accurate spin-dependent electron liquid correlation energies for local spin density calculations: A critical analysis. Canadian Journal of Physics. 1980;58(8):1200–1211. [Google Scholar]
- 59.Hay PJ. Gaussian basis sets for molecular calculations. The representation of 3d orbitals in transition-metal atoms. The Journal of Chemical Physics. 1977;66(10):4377–4384. [Google Scholar]
- 60.McGrath MP, Radom L. Extension of Gaussian-1 (G1) theory to brominecontaining molecules. The Journal of Chemical Physics. 1991;94(1):511–516. [Google Scholar]
- 61.McLean AD, Chandler GS. Contracted Gaussian basis sets for molecular calculations. I. Second row atoms, Z=11–18. The Journal of Chemical Physics. 1980;72(10):5639–5648. [Google Scholar]
- 62.Wachters AJH. Gaussian basis set for molecular wavefunctions containing third-row atoms. The Journal of Chemical Physics. 1970;52(3):1033–1036. [Google Scholar]
- 63.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Montgomery JA, Peralta JE, Ogliaro F, Bearpark M, Heyd JJ, Brothers E, Kudin KN, Staroverov VN, Kobayashi R, Normand J, Raghavachari K, Rendell A, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam JM, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas, Foresman JB, Ortiz JV, Cioslowski J, Fox DJ Gaussian 09, Revision B.01. Wallingford CT: 2009. [Google Scholar]
- 64.Singh N, Warshel A. Towards accurate microscopic calculation of solvation entropies: Extending the restraint release approach to studies of solvation effects. Journal of Physical Chemistry B. 2009;113(20):7372–7382. doi: 10.1021/jp811063v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Singh N, Warshel A. Absolute binding free energy calculations: On the accuracy of computational scoring of protein-ligand interactions. Proteins-Structure Function and Bioinformatics. 2010;78(7):1705–1723. doi: 10.1002/prot.22687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Strajbl M, Sham YY, Villa J, Chu ZT, Warshel A. Calculations of activation entropies of chemical reactions in solution. Journal of Physical Chemistry B. 2000;104(18):4578–4584. [Google Scholar]
- 67.Dove MT. Introduction to the theory of lattice dynamics. Press CU; 1993. editor. [Google Scholar]
- 68.Guthrie JP. Hydration of carbonyl compounds, an analysis in terms of multidimensional marcus theory. Journal of the American Chemical Society. 2000;122(23):5529–5538. [Google Scholar]
- 69.Wang H. An accurate theoretical study of energy barriers of alkaline hydrolysis of carboxylic esters. Res Chem Intermed. 2012;38(9):2175–2190. [Google Scholar]
- 70.Cahn RS, Ingold C, Prelog V. Specification of molecular chirality. Angewandte Chemie International Edition in English. 1966;5(4):385–415. [Google Scholar]
- 71.Prelog V, Helmchen G. Basic principles of the CIP-system and proposals for a revision. Angewandte Chemie International Edition in English. 1982;21(8):567–583. [Google Scholar]
- 72.Hwang JK, Warshel A. Semiquantitative calculations of catalytic free-energies in genetically modified enzymes. Biochemistry. 1987;26(10):2669–2673. doi: 10.1021/bi00384a003. [DOI] [PubMed] [Google Scholar]
- 73.Singh N, Warshel A. A comprehensive examination of the contributions to the binding entropy of protein–ligand complexes. Proteins: Structure, Function, and Bioinformatics. 2010;78(7):1724–1735. doi: 10.1002/prot.22689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reetz MT, Krebs GPL. Challenges in the directed evolution of stereoselective enzymes for use in organic chemistry. Comptes Rendus Chimie. 2011;14(9):811–818. [Google Scholar]
- 75.Lee S-C, Kim J-H, Kim H-S. Design and evolution of biocatalysts. Current Organic Chemistry. 2010;14(17):1894–1901. [Google Scholar]
- 76.Kazlauskas R. Engineering enantioselectivity in enzyme-catalyzed reactions. Wiley-VCH Verlag GmbH & Co, KGaA: Protein Engineering Handbook; 2008. pp. 15–46. [Google Scholar]
- 77.Reetz MT. Directed evolution of enantioselective enzymes: An unconventional approach to asymmetric catalysis in organic chemistry. The Journal of Organic Chemistry. 2009;74(16):5767–5778. doi: 10.1021/jo901046k. [DOI] [PubMed] [Google Scholar]
- 78.Morley KL, Kazlauskas RJ. Improving enzyme properties: when are closer mutations better? Trends in Biotechnology. 2005;23(5):231–237. doi: 10.1016/j.tibtech.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 79.Pan J, Yu H-L, Xu J-H, Lin G-Q. Advances in biocatalysis: Enzymatic reactions and their applications. In: Ma S, editor. Asymmetric Catalysis from a Chinese Perspective. Volume 36. Springer Berlin Heidelberg: Topics in Organometallic Chemistry; 2011. pp. 67–103. [Google Scholar]
- 80.Gröger H. Catalytic Asymmetric Synthesis. John Wiley & Sons, Inc.; 2010. Enzyme-catalyzed asymmetric synthesis; pp. 269–341. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.