Abstract
Cattle have a limited range of immunoglobulin genes which are further diversified by antigen independent somatic hypermutation in fetuses. Junctional diversity generated during somatic recombination contributes to antibody diversity but its relative significance has not been comprehensively studied. We have investigated the importance of terminal deoxynucleotidyl transferase (TdT) -mediated junctional diversity to the bovine immunoglobulin repertoire. We also searched for new bovine heavy chain diversity (IGHD) genes as the information of the germline sequences is essential to define the junctional boundaries between gene segments. New heavy chain variable genes (IGHV) were explored to address the gene usage in the fetal recombinations. Our bioinformatics search revealed five new IGHD genes, which included the longest IGHD reported so far, 154 bp. By genomic sequencing we found 26 new IGHV sequences that represent potentially new IGHV genes or allelic variants. Sequence analysis of immunoglobulin heavy chain cDNA libraries of fetal bone marrow, ileum and spleen showed 0 to 36 nontemplated N-nucleotide additions between variable, diversity and joining genes. A maximum of 8 N nucleotides were also identified in the light chains. The junctional base profile was biased towards A and T nucleotide additions (64% in heavy chain VD, 52% in heavy chain DJ and 61% in light chain VJ junctions) in contrast to the high G/C content which is usually observed in mice. Sequence analysis also revealed extensive exonuclease activity, providing additional diversity. B-lymphocyte specific TdT expression was detected in bovine fetal bone marrow by reverse transcription-qPCR and immunofluorescence. These results suggest that TdT-mediated junctional diversity and exonuclease activity contribute significantly to the size of the cattle preimmune antibody repertoire already in the fetal period.
Introduction
Somatic recombination generates a large immunoglobulin repertoire by the assembly of variable (V), diversity (D) and joining (J) gene segments coding for heavy chains and V and J segments coding for light chains [1]. In cattle and several other domestic animals the germline population of V, D and J segments is too small to provide sufficient immunoglobulin diversity. These species use additional mechanisms in order to expand the preimmune repertoire, which is the repertoire in use before exposure to environmental antigens [2]. Long immunoglobulin heavy chain D genes are characteristic of bovine immunoglobulins as they contribute to the exceptionally long third complementarity determining regions of the heavy chains (CDR3H) [3]–[5]. We have previously shown that somatic hypermutation (SHM) diversifies the immunoglobulin repertoire by introducing mutations especially in the CDR3H region, already at the fetal period, before the exposure to external antigens [6]. In addition to SHM, terminal deoxynucleotidyl transferase (TdT) mediated junctional diversity has been reported in cattle but its significance to the preimmune repertoire has not been thoroughly investigated [7].
TdT adds nontemplated (N) nucleotides to the single-strand DNA ends, in connection with V(D)J recombination which is guided by recombination signal sequences (RSSs). These conserved sequences flank each V, D and J segment. [1]. The recombination process requires multiple enzymes such as polymerases, nucleases and ligases. A complex encoded by recombination-activating genes (RAG1 and RAG2) plays a crucial role in bringing the two RSSs together and cleaving the double stranded DNA. As a result, the cleaved free end forms a DNA hairpin which is then opened by the Artemis:DNA-dependent protein kinase (DNA-PK) nuclease complex at a random site. Sometimes the cleavage generates palindromic (P) nucleotides [8]. Whenever TdT is present and active in the cell, it increases the variability of the junctions by adding N nucleotides to the available 3′-OH ends adjacent to the P nucleotides. Also excision of nucleotides by largely uncharacterized exonucleases occurs [9]. As the N- and P-nucleotide additions are largely random they often result in nonproductive rearrangements [10], [11]. In mice, the length of productive N additions is 2–5 bp in vivo. In vitro experiments have shown that TdT is capable of catalyzing even longer than 1 kb nucleotide additions [12] with a bias towards dGMP residues [13]. In addition to rearranged immunoglobulin genes, N additions also take place in genes encoding T-cell receptors [14].
TdT belongs to the PolX family of DNA polymerases with Polβ, Polλ and Polµ in eukaryotes [15]. It is considered the only canonical template independent DNA polymerase, although Polµ has also been reported to have template independent functions [16]. In mammals, alternative splicing generates two or three TdT isoforms among which functional differences have been observed. In mouse two isoforms, mTdTS and mTdTL have been identified [17]. All of the murine isoforms are expressed after birth and N additions are usually found only in rearranged IGH genes. The function of mTdTL still remains unclear. It is suggested that rather than adding nucleotides it may function as an exonuclease, trimming the coding ends of V, D and J segments [18], [19]. Human and cattle have three isoforms: TdTS, TdTL1 and TdTL2 [20], [21]. In humans, both of the long isoforms possess 3′→5′ exonuclease activity. Human TdTS, on the contrary, may carry out nucleotide elongation during V(D)J recombination. The human TdTs are expressed already in fetal life in T- and B-cell progenitors in thymus and bone marrow [21].
In this study, we first complemented the current IGH gene repertoire by searching new immunoglobulin variable (IGHV) and diversity (IGHD) genes. Accurate reference germline sequences were a prerequisite for analysing the junctional boundaries in fetal recombinations. Junctional diversity was then analysed from fetal cDNA libraries of both heavy and light chains. Furthermore, the expression of TdT and its splice variants was investigated by reverse transcription (RT) qPCR and triple-colour immunofluorescence in several tissues. We focused on fetal recombinations as in cattle de novo B lymphopoiesis takes place in fetal bone marrow and lymph nodes, as indicated by expression of pre-B cell markers [22], [23]. Our results indicate significant TdT-induced junctional diversity in bovine immunoglobulins and suggest a novel diversification mechanism which involves extensive trimming of IGHD genes.
Materials and Methods
Ethical statement
Fetal and adult tissue samples were collected from a local abattoir, where cattle are slaughtered on a daily basis and used for human consumption. We took our samples from slaughtered, healthy animals. No experimental procedures were done on living animals and they were not euthanized for this study. Therefore, no ethical permit was required.
Cloning and sequencing of bovine germline IGHV genes
Germline IGHV genes were cloned and sequenced as previously described [6]. Skeletal muscle genomic DNA was extracted from three fetuses, (aged 182, 240 and 270 gestation days (gd)) and a 51-days-old calf. The reads of an IGHV were considered reliable if the same sequence was identified at least twice. The new IGHV sequences are presented in Table S1.
Extraction of bovine IGHD sequences from genomic sequencing data
Available bovine genomic sequencing data (including the NCBI Unfinished high throughput genomic sequences and the NCBI trace archive) was explored for previously unidentified IGHD genes using the fuzznuc motif search [24] for consensus D-RSS sequences. The PROSITE search motives used were GGTTTTTGT N(11,13)CACNGTGN(6,160)CACNGTGN(11,13)ACAAAAACC with up to 4 mismatches and GG[TA]TTN[ATG][GA][ATG]N(12)N[AG][TC]NGT[GC] N(30,180) CAC[ACT][AG][TC][GA] N(12)NC[AC][ACG]AAA[ACG][CT] with up to 2 mismatches. The known and newly identified IGHD genes are presented in the Table S2.
Preparation of fetal IGH, IGL and IGK cDNA libraries
For the heavy chain library, samples of ileum, spleen and bone marrow were collected from fetuses of 240 and 270 gd. First-strand cDNA was synthesised using SuperScript III First-Strand Synthesis SuperMix according to manufacturer's instructions. First-strand cDNA and the IGH cDNA library was prepared as described in [6].
For the light chain libraries, total RNA was purified from ileum and bone marrow of 270 gd old fetus. The first-strand cDNA was primed using equal amounts of oligo (dT)20 and random hexamer primers and SuperScript III First-Strand Synthesis SuperMix was used for cDNA synthesis. Furthermore, the variable and joining segments were amplified with PCR using Phusion MasterMix (Fermentas). The PCR reaction contained 1 x Phusion MasterMix, 0.5 µM forward and reverse primers (IgLg1fwd and IgLg1 rev for lambda light chains and IgKg2 fwd and IgKg2 rev for kappa light chains, Table 1) and 0.5 µl of the cDNA template (ileum or bone marrow). Two-step PCR protocol cycling conditions consisted of an initial denaturation of 98°C for 30 s, followed by 29 cycles of 98°C for 10 s, 72°C for 15 s, and a final extension of 72°C for 7 min. PCR products were then electrophoresed and purified. Approximately 20 ng of the purified PCR product was ligated into the pCR Blunt II-TOPO Vector and transformed into TOP10 E. coli (Life technologies). The TOP10 E. coli were grown overnight at 37°C on LB-kanamycin (50 µg/ml) plates. A total number of 48 single colonies were picked up, purified and sequenced by GATC Biotech AG (Konstanz, Germany).
Table 1. PCR primers and probes.
| Primer | Sequence 5′-3′ |
| TdT-FW | GCTTCAGGTACAGAACATA |
| TdT-REV | GTCTGTTCTCACAACAAG |
| TdT probe | FAM-ACT[+C]CT[+T]GA[+T]GT[+C]TCCTG-BHQ1* |
| T7 | TAATACGACTCACTATAGGG |
| T3 | ATTAACCCTCACTAAAGGGA |
| TdT_tr1_1925_FW | AGACCAAGTGCACATATG |
| TdT_tr1_1925_PR | FAM-ATTTCTTCTTCACTTTCCGCTTTGAGA-BHQ1 |
| TdT_tr1_1925_RV | GGTTCAATGTAGTCCAATC |
| TdT_tr2_131_FW | TGGTCAGGTTTTGGATTTC |
| TdT_tr2_131_PR | HEX-CAGAAATGCCCACACAGCCTC-BHQ1 |
| TdT_tr2_131_RV | CCTGTCATGGTGACAAAG |
| 18S fwd | TGGTTGCAAAGCTGAAACTTAAAG |
| 18S probe | HEX-CCTGGTGGTGCCCTTCCGTCA-BHQ1 |
| 18S rev | AGTCAAATTAAGCCGCAGGC |
| IgLg1fwd | GGCCCAGGCTGTGCTGACTC |
| IgLg1 rev | TGATGGTGCTGCCGTCTGCC |
| IgKg2 fwd | TGTGCTGACCCAGACTCCCCT |
| IgKg2 rev | ACAGTTCCGGTCTTCAGCTGCTC |
| IgH fwd1 | TTGTGCTSTCAGCCCCCAGA |
| IgH rev1 | CGCAGGACACCAGGGGGAAG |
| IGHV fwd3 | GGACAACTCCAAGAGCCAAG |
| IGHJrev2-FAM | TGAGGAGACGGTGACCMKGAG |
*Nucleotides in square brackets refer to locked nucleic acids.
Spectratyping
Total RNA was extracted as in the previous sections. The first-strand cDNA was reverse transcribed using RevertAid Premium Reverse Transcriptase (Fermentas) and primed with equal amounts of oligo (dT)20 and random hexamer primers. First-strand cDNA was used as a template for the nested PCR. For the first round primers IgH fwd1 and IgH rev1 were used, amplifying the region from leader1 exon to the CH1 region. This PCR product was used as a template for the second round PCR. Here, primers covering the CDR3H region were used (IGHV fwd3 and IGHJ rev2-FAM, Table 1). Capillary electrophoresis was run at the Sequencing unit of the Institute of Biotechnology (University of Helsinki). The raw data were analysed in PeakScanner (ABI). Data was filtered and combined from 24 samples (four fetuses, six tissues: bone marrow, ileum, liver, lymph node, spleen, and thymus), and the signal density function with Gaussian smoothing kernel was computed in R [25].
Sequence analysis of V(D)J junctions
The sequence data from cDNA libraries were analysed with Geneious Pro software version 6.0 (Biomatters, New Zealand), the EMBOSS package [24], MUSCLE version 3.7 [26], and R software [25]. Sequences were discarded when they did not cover the entire CDR3 region. Sequences were aligned with previously detected germline sequences [3], [27]–[30] (see also Tables S1, S2 and S3) using Smith-Waterman local alignment algorithm implemented in Biostrings R-package [31] and its heuristic approximation implemented in blastn [32]. The heavy chain cDNA sequences were aligned against custom bovine-specific IGHV, IGHD and IGHJ gene databases by the pairwiseAligment function in Biostrings using the following parameters: match = 1, mismatch = -1, gapOpening = -4, gapExtension = -5 (for variable and joining genes) or -0.3 (diversity genes), and type of aligment = local. The boundaries corresponding to the V, D and J segments derived from specific IGHV, IGHD and IGHJ germline sequences were determined from the coordinates of the best pairwise alignment with the query sequence. The boundaries for V and J segments were first determined and the subsequence between these coordinates was then used for querying the IGHD database (Table S4). The gap extension penalty was optimized to a low level (-0.3) to extend the alignments, since the IGHD sequences contain shared repetitive short motifs (Table S2). Boundaries for overlapping V, D and J segments were set at the middle of the overlap. The light chain sequences were analysed in a similar way except that blastn was used for pairwise alignments.
Recombination-associated exonuclease activity was determined for each end of the germline reference gene acting as the donor sequence (Table S5). For this, we counted the number of donor sequence nucleotides excluded from the recombined sequence. When the donor sequence end had not been modified, potential P nucleotides complementary to the donor sequence end were identified. The reverse complement of the donor sequence end was compared nucleotide by nucleotide with the recombined query gene sequence in the VD or DJ junction. The remaining nucleotides in the VD or DJ junction were classified as N nucleotides (Tables S4 to S7).
Assessment of the expression of TdT splice variants by reverse transcription-qPCR (RT-qPCR)
Tissue samples from 6 fetal and 5 adult cattle were collected from a local abattoir, snap frozen in liquid nitrogen, and stored at −80°C. Total RNA was extracted from liver, ileum, spleen, lymph node, thymus and bone marrow using Eurozol (EuroClone, Italy) as described in [6].
Reverse transcription into cDNA was performed using 1 µg of total RNA with Revert-AID M-MuLV Reverse Transcriptase (Fermentas, Germany) or SuperScript III First-Strand Synthesis SuperMix (Life technologies, Germany). First-strand cDNA was primed with oligo(dT)20 (Oligomer, Finland) and synthesis was performed according to manufacturer's instructions.
Three sets of primers and probes were designed for the different splice variants (Figure 1 and Table 1). TdT-FW and TdT-REV recognize all three forms of the bovine TdT mRNA. TdT_tr_1_1925_FW and TdT_tr_1_1925_RV recognize the splice variant II and the TdT_tr2_131_FW and TdT_tr2_131_RV recognize the splice variant I. All primers and probes were designed by Sigma-Aldrich. Amplification was carried out using the Stratagene Mx3005P real-time PCR system (Agilent Technologies, USA). Cycling conditions were: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, 60°C for 30 s and 72°C for 30 s. Reactions were performed in duplicates.
Figure 1. qPCR primers and probes for bovine TdT splice variants.
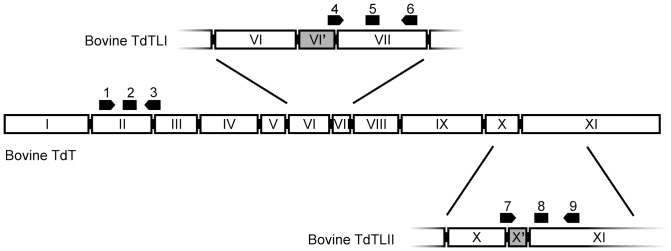
Numbering of exons according to human TdT [68]. Extra exons in the splice variants (VI’ and X’) are numbered according to [20]. Black arrows indicate the primers and rectangles the probes for qPCR, as specified in Table 1. 1. TdT-FW, 2. TdT probe, 3. TdT-REV, 4. TdT_tr2_131_FW, 5. TdT_tr2_131_PR, 6. TdT_tr2_131_RV, 7. TdT_tr1_1925_FW, 8. TdT_tr1_1925_PR, 9. TdT_tr1_1925_RV.
TdT expression was quantified using RT-qPCR. From each tissue the threshold cycle for TdT was normalized with that of 18S RNA. In order to compare the relative changes in the splice variant I and II gene expression, the 2−ΔΔCt method was used [33]. The 18S-normalized values were calibrated with the normalized value of the expression in the adult thymus. The range for relative expression was between 0 (no detectable expression) and 1 (the same level of expression as in the calibrator thymus). The statistical analysis was done in R with non-parametric Friedman two-way ANOVA followed by pair-wise comparison using Wilcoxon-Nemenyi-McDonald-Thompson test when assessing the all-over TdT mRNA expression. Kruskal-Wallis ANOVA followed by pair-wise comparison using Nemenyi-Damico-Wolfe-Dunn test was used with splice variant analysis [25], [34].
Triple immunofluorescence and image analysis
Tissue sections were deparaffinized, subjected to heat-induced antigen retrieval in 10 mM Tris-HCl pH 9.5, 1 mM EDTA pH 8, and blocked with 1% goat serum. They were then incubated in a mixture of a rabbit polyclonal anti- bovine TdT antibody (Dako, Denmark) and the rat monoclonal anti-CD3 antibody CD3-12 (Santa Cruz Biotechnology, TX), washed, incubated in a mixture of a goat anti-rabbit Ig Alexa647 antibody (Dako) and a goat anti-rat Ig DyLight488 antibody (Jackson Immunoresearch, PA), washed again, incubated in the monoclonal mouse anti-human CD79α antibody HM57 (Dako), washed and finally incubated in a donkey anti-mouse Ig DyLight549 antibody preadsorbed against rat Igs (Jackson Immunoresearch). The sections were counterstained with DAPI, fixation autofluorescence was suppressed by incubation in 0.1% Sudan Black B in 70% ethanol, and the coverslips were mounted using Dako Immunofluorescence Mounting Medium.
The stained sections were photographed in the four fluorescence channels using a Zeiss AxioVision microscope. To assess the phenotypes of the TdT+ cells, photomicrographs of randomly selected TdT+ cells were observed in the CD3 and CD79α channels. CD79α+CD3− cells were counted as B lymphocytes and all CD3+ cells as T lymphocytes. To calculate the proportions of TdT+ cells among all B lymphocytes, TdT+CD79α+CD3− cells were first manually counted in randomly selected B-cell rich areas in tissue sections (a minimum of five 0.15 mm2 image fields produced with a 20× objective per tissue per animal). The total numbers of CD79α+ cells in the areas analyzed were then estimated by dividing the total area of CD79α+ immunofluorescence with the average area in a single B lymphocyte in threshold-segmented images, using ImageJ [35]. The minimum number of B cells thus analysed per animal was 80 in fetal bone marrow, 2440 in lymph nodes, 900 in spleen, 1280 in fetal liver and 21500 in fetal Peyer's patch, reflecting the B cell densities in these tissues. For the tissues where no TdT+ B cells were observed, a minimum of 1200 B cells were screened per section (except for adult bone marrow and liver, where B cells are very rare).
Results
Data mining and sequencing uncovers potentially novel IGHV and IGHD genes
In order to ensure a complete set of reference germline immunoglobulin heavy chain gene sequences for analyses of junctional diversity, we sequenced the IGHV genes in the four animals used in this study and mined all available bovine genomic sequence data for IGHD genes.
In addition to the 10 functional IGHV genes previously annotated in the genomic data [36], we identified 26 new germline IGHV sequences. These were assigned temporary gene designations based on the IMGT nomenclature [37]. The 26 sequences have been deposited to GenBank as KJ491073-KJ491098 and are listed in Table S1. They all belong to the subgroup IGHV1 and include potentially new genes as well as new allelic variants of existing genes.
Data mining uncovered four new IGHD sequences (IGHDS10 to IGHDS13, Table S2) in addition to the previously characterized 10 IGHD genes [7], [28]. Pairwise alignments between the new IGHD sequences and immunoglobulin cDNAs strongly suggested the presence of a fifth novel germline sequence IGHDS14 that was related to IGHDS12 (Figure S1). To deduce the IGHDS14 sequence, multiple sequence alignment was done among the corresponding cDNAs that were derived from at least 20 different recombinations based on variable IGHV gene usage and CDR3H length. The consensus sequence representing the partial IGHDS14 sequence was then determined (Figure S2, Table S2). The five novel IGHD sequences represent uncharacterized genes or allelic variants of existing genes. Their length ranged from 31 bp (IGHDS10) to 154 bp (IGHDS12), the longest of bovine IGHD genes identified to date. They were variably used in immunoglobulin gene recombinations (Table 2).
Table 2. Expressed combinations of IGHV, IGHD and IGHJ segments in bovine fetal bone marrow, ileum and spleen.
| V gene | IGHDS5 | IGHDS3 | IGHDS7 | IGHDS8 | IGHDS2 | IGHDS14 | IGHDS4 | IGHDS11 | IGHDS12 | IGHDS9 | IGHDS1 | IGHDS10 | IGHDS13 | IGHDS6 | sum |
| IGHV1S3 | 31/0/2 | 19/0/0 | 24/0/1 | 16/0/13 | 3/0/5 | 8/0/0 | 11/0/0 | 4/0/0 | 0/0/0 | 1/0/0 | 3/0/0 | 1/0/0 | 0/0/0 | 1/0/0 | 143 |
| IGHV1S39 | 84/0/0 | 2/0/0 | 8/0/1 | 0/0/3 | 1/0/0 | 1/0/0 | 0/0/0 | 2/0/0 | 1/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 103 |
| IGHV1S15 | 25/0/0 | 32/0/0 | 5/0/0 | 9/0/0 | 13/0/0 | 0/0/0 | 7/0/0 | 1/0/0 | 0/0/0 | 2/0/0 | 1/0/0 | 1/0/0 | 3/0/0 | 0/0/0 | 99 |
| IGHV1S28 | 27/0/0 | 8/0/0 | 10/0/0 | 2/0/0 | 10/0/0 | 2/0/0 | 3/0/0 | 0/0/0 | 1/0/0 | 0/0/1 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 64 |
| IGHV1S1 | 14/0/0 | 3/0/1 | 3/0/0 | 2/0/9 | 2/0/1 | 17/0/0 | 0/0/0 | 0/0/0 | 5/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 57 |
| IGHV1S6 | 15/0/0 | 3/0/0 | 4/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 1/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 23 |
| IGHV1S18 | 15/0/0 | 4/0/0 | 2/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 21 |
| IGHV1S34 | 11/0/0 | 6/0/0 | 1/0/0 | 1/0/0 | 1/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 1/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 21 |
| IGHV1S11 | 6/0/0 | 3/0/2 | 4/0/0 | 0/0/1 | 0/0/0 | 3/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 1/0/0 | 0/0/0 | 0/0/0 | 20 |
| IGHV1S14 | 2/0/0 | 1/0/0 | 5/0/0 | 2/0/3 | 1/0/0 | 1/0/0 | 1/0/0 | 0/0/0 | 0/0/0 | 1/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 17 |
| IGHV1S10 | 8/0/0 | 2/0/0 | 3/0/0 | 0/0/0 | 1/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 1/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 15 |
| IGHV1S33 | 6/0/0 | 1/0/0 | 4/0/0 | 1/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 1/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 13 |
| IGHV1S31 | 9/0/0 | 1/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 1/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 11 |
| IGHV1S16 | 4/0/0 | 0/0/0 | 1/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 5 |
| IGHV1S2 | 2/0/0 | 1/0/0 | 0/0/0 | 0/0/2 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 5 |
| IGHV1S24 | 0/1/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 4/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 5 |
| IGHV1S40 | 3/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 2/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 5 |
| IGHV1S4 | 0/0/0 | 1/0/0 | 0/0/0 | 0/0/3 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 4 |
| IGHV1S35 | 1/0/0 | 0/0/0 | 1/0/0 | 0/0/0 | 1/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 3 |
| IGHV1S7 | 1/0/0 | 1/0/0 | 0/0/0 | 1/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 3 |
| IGHV1S26 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/1 | 0/0/0 | 0/0/0 | 1/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 2 |
| IGHV1S29 | 1/0/0 | 0/0/0 | 1/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 2 |
| IGHV1S19 | 0/0/0 | 0/0/0 | 1/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 1 |
| IGHV1S27 | 0/0/0 | 0/0/0 | 0/0/0 | 1/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 1 |
| IGHV1S36 | 0/0/0 | 1/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 1 |
| IGHV1S38 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/1 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 1 |
| per JH1/2/6 | 265/1/2 | 89/0/3 | 77/0/2 | 35/0/36 | 35/0/6 | 32/0/0 | 24/0/0 | 9/0/0 | 9/0/0 | 8/0/1 | 4/0/0 | 3/0/0 | 3/0/0 | 1/0/0 | 594/1/50 |
| Sum | 268 | 92 | 79 | 71 | 41(*) | 32 | 24 | 9 | 9 | 9 | 4 | 3 | 3 | 1 | 645 |
A total of 645 cDNA sequences were analyzed from bone marrow, ileum and spleen. Each cell shows the numbers of cDNAs containing the JH1, JH2 and JH6 gene segment. The IGHV segment is specified by the row and the IGHD gene by the column. As an example, the top left cell shows that there were 31 cDNAs containing the combination IGHV1S3-IGHDS5-JH1 and 2 cDNAs corresponding to IGHV1S3-IGHDS5-JH6. The most commonly expressed genes were IGHV1S3, IGHDS5 and JH1. The most common combination was IGHV1S39-IGHDS5-JH1. (*) In 31 cDNAs, the ends of IGHDS2 were extensively trimmed (Table 6).
A limited range of gene combinations is found in the immunoglobulin cDNAs
cDNA libraries from fetal bone marrow (specific for IGH, IGL, IGK), ileum (IGH, IGL, IGK) and spleen (IGH) were analyzed for various combinations of immunoglobulin variable, diversity and joining genes (Tables 2 and 3). Twenty-six IGHV genes were found in the cDNA sequences (N = 645) of which five genes (IGHV1S3, IGHV1S39, IGHV1S15, IGHV1S28 and IGHV1S1) accounted for 72%. Thirteen IGHD genes were detected in the cDNA sequences. IGHDS5 (DH5 [21]) was used in 42% and IGHJS1 (JH1 [27]) in 92% of the sequences. The most common combination was IGHV1S39-IGHDS5-IGHJS1, which occurred in 13% of all cDNAs analyzed. The long IGHDS2 (148 bp), IGHDS12 (154 bp) and IGHDS14 (119 bp or longer) genes were found in 13% of the recombinations.
Table 3. Expressed combinations of IGLV and IGLJ gene segments in bovine fetal bone marrow and ileum.
| IGLJ3 | IGLJ2 | |
| IGLV30 | 20 | 3 |
| IGLV39 | 7 | 1 |
| IGLV49 | 7 | |
| IGLV8 | 6 | 1 |
| IGLV43 | 4 | |
| IGLV28 | 4 | |
| IGLV25 | 3 | |
| IGLV55 | 2 | |
| IGLV35 | 2 | |
| IGLV2 | 2 | |
| IGLV6 | 2 | |
| IGLV56 | 1 |
n = 65.
The immunoglobulin λ cDNA sequences from bone marrow and ileum of a 270 days old fetus matched to 12 of the previously identified 25 potentially functional IGLV genes [30]. All of these genes belong to subgroup 1. The preferential gene usage did not differ between the two tissues. IGLV30 was the most common of the variable genes expressed, and the combination IGLV30-IGLJ3 accounted for 35% of the cDNA sequences (Table 3). The second common variable gene used was IGLV39 (12%). IGLJ2 was used only in 9% of sequences whereas IGLJ3 was used in 91%. We identified the expression of 3 κ variable genes out of the 8 potentially functional genes [30]. IGKV19 was used in 64% (Table 4) whereas 35% of the sequences contained IGKV10. Also IGKV17 was detected in 1% of the sequences. In 97% of the sequences, IGKJ1 was used.
Table 4. Expressed combinations of IGKV and IGKJ gene segments in bovine fetal bone marrow and ileum.
| IGKJ1 | IGKJ2 | |
| IGKV19 | 53 | |
| IGKV10 | 28 | 1 |
| IGKV17 | 1 |
n = 83.
N nucleotide additions and exonuclease activity shape the CDR3H region
We analyzed the CDR3H encoding region in 645 cDNA sequences derived from bone marrow, ileum and spleen of two nearly full term fetuses (Tables S4 and S5). The average length of CDR3H encoding region in the recovered clones was 74.9 nucleotides. In 8.4% of the sequences the CDR3H encoding region was over 100 bp long suggesting a second subpopulation of bovine IGH cDNAs with long CDR3H encoding region [3]. This was confirmed by a separate spectratyping assessment of the CDR3H lengths of fetal thymus, spleen, ileum, lymph node, liver and bone marrow (Figure 2).
Figure 2. Bovine fetal IGH spectratype.
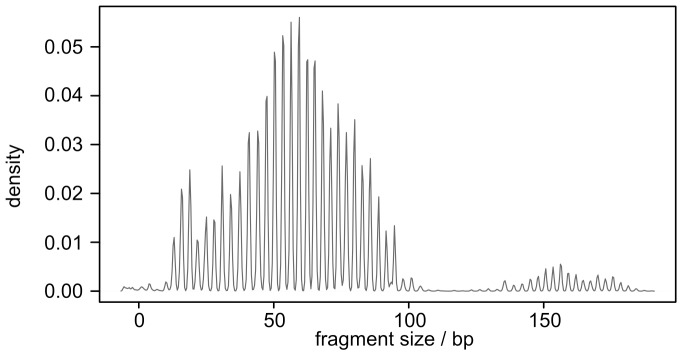
Pooled data from 24 samples (four fetuses, six tissues/fetus: thymus, spleen, ileum, lymph node, liver, and bone marrow). Fragment size corresponds to the length of CDR3.
The median of N nucleotides was 1 in VD and 2 in DJ junctions (n = 645, Figure 3). They were found in 65% in VD and 68% in DJ junctions. All in all, 90% of the sequences contained N nucleotides. Some extremely long N additions could also be seen. More than 10 N additions (range 10 to 36 nt) were found in 4.5% of VD junctions and 3.6% (range 10 to 16 nt) of DJ junctions. Palindromic P nucleotides were also seen, with 16% of VD junctions (range 1 to 6) and 18% of DJ junctions (range 1 to 3) showing P additions (Figure 3 and Table 5). The base profile of N nucleotide additions in VD junctions was dominated by T (33%) and A (31%) followed by G (19%) and C (17%). The profile was more homogenous in the DJ junctions with about equal frequency of T and A (26% each) vs. G (28%) and C (21%). We could not detect conserved short nucleotide sequences (CSNS) that have previously been reported in adult bovine VDJ recombinations [7].
Figure 3. Junctional diversity in bovine fetuses.
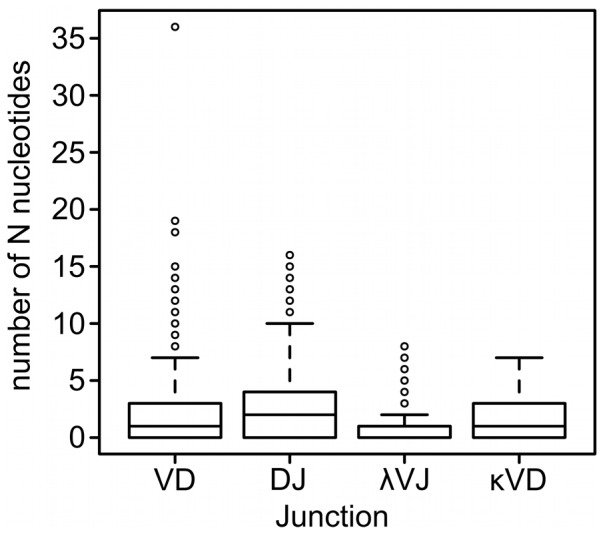
N nucleotide additions in IGH (VD and DJ junctions, n = 645) IGL (λVJ junction, n = 65) and IGK (κVJ junction, n = 83). Dark line in the middle represents the median and 50% of the cases lie within the box. Whiskers extend to 1.5 times the height of the box. Circles represent outliers. We did not detect statistically significant differences between tissues or individuals. Heavy chain data is pooled from bone marrow, ileum and spleen (two fetuses). Light chain data is pooled from bone marrow and ileum (one fetus).
Table 5. Analysis of nucleotide additions in bovine fetal IGH (bone marrow), IGL and IGK (bone marrow and ileum).
| Junction | Number of sequences | Median number of N nucleotides (range) | Sequences with N additions (%) | Long (>10) N additions (%) | Median number of P nucleotides (range) | Sequences with P additions (%) |
| VD | 645 | 1 (0–36) | 65 | 4.5 | 0 (0–6) | 16 |
| DJ | 645 | 2 (0–16) | 68 | 3.6 | 0 (0–3) | 18 |
| VJ λ | 65 | 0 (0–8) | 36.9 | 0 | 0 (0–1) | 2 |
| VJ κ | 83 | 1 (0–7) | 60 | 0 | 0 (0–1) | 2 |
As the readout for the exonuclease activity, we used the loss of nucleotides from the ends of V, D, and J gene segments. The exonuclease activity removed a median of 2 nucleotides from the 3′ end of IGHV and from the 5′ end of IGHJ. We also detected extensive trimming of IGHD gene ends. The median value of the number of deleted nucleotides from the ends of IGHD genes was 5 in a VD junction and 6 in a DJ junction. There was a statistically significant difference between the number of deleted nucleotides from D genes vs. V or J genes (Mann-Whitney U test, P<1e-16).
N-nucleotide additions occur in λ and κ light chains but to a lesser extent than in the heavy chains
We sequenced 65 IGL and 83 IGK cDNA clones from bone marrow and ileum of the same fetuses as above. The numbers of N additions found in VJ junctions were similar between the two tissues. Therefore, we pooled the results from bone marrow and ileum. Nontemplated additions were found in 36.9% (24) of IGL clones and 60% (50) of IGK clones. The median of N nucleotides was 0 in IGL and 1 in IGK (Figure 3). There was a statistically significant difference between the number of N additions in IGL and IGK (Mann-Whitney U test; P = 0.006). Very few P nucleotides could be detected in light chains (Table 5). Exonuclease activity was also detected in light chains: a median value of 2.5 nucleotides was excised from the 3′ end of IGLV and IGKV genes. The corresponding numbers in the 5′ end of joining genes were as follows: in IGLJ 1 bp and in IGKJ 3 bp (Mann-Whitney U test P<7e-08). As in the heavy chain sequences, the junctional base profile was dominated by T (36.4%) and A (24.3%) followed by C (20.4%) and G (19.0%).
TdT and its splice variants are expressed in bone marrow in bovine fetuses
The expression of TdT mRNA was measured with RT-qPCR in 3 fetal and 2 adult cattle. The general primers located to exon 2 and can also amplify the long isoforms (Figure 1). Thymus, bone marrow and lymph node showed elevated expression levels compared to liver, ileum and spleen (Figure 4, P = 0.003, α = 0.05). Fetuses did not differ from adults.
Figure 4. TdT mRNA expression level in adults and fetuses.
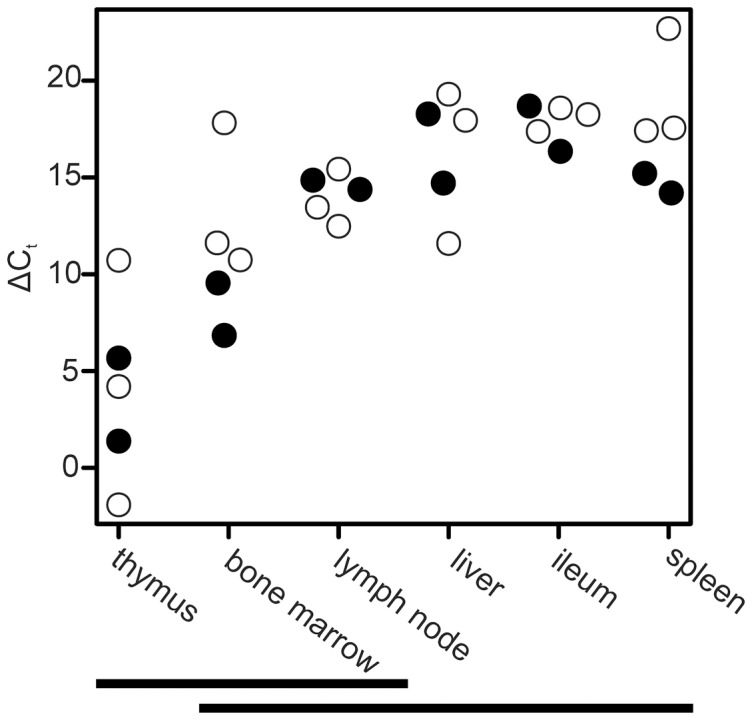
The expression level was measured with RT-qPCR. Material consisted of 3 fetuses and 2 adults. Thymus, bone marrow and lymph node had elevated expression levels compared to liver, ileum and spleen. Fetuses did not differ from adults. 18S normalized cycle threshold values (ΔCt) are shown. Tissues not differing statistically (α = 0.05) from each other are indicated by a horizontal bar. White points indicate fetuses and black points indicate adults.
Expression of known TdT isoforms, bovineTdTL1 and bovineTdTL2, was also assessed with RT-qPCR. The long isoforms L1 and L2 both contain an extra exon VI’ and X’ respectively (Figure 1). The highest expression levels were seen in thymus. In fetuses, the expression of both L1 and L2 differed between thymus and spleen and between thymus and ileum (P<0.0004, α = 0.01, data not shown). There were no statistical differences between the levels of tissue specific expression of either long isoform in adults (data not shown).
To identify the cell types expressing TdT in various tissues, we performed triple immunofluorescence for TdT, the B lymphocyte marker CD79α and the T lymphocyte marker CD3. In the fetal bone marrow, 41±13% of the TdT positive cells were identified as CD79α+CD3− B lymphocytes and 8.2±2.9% as CD3+ T lymphocytes (n = 5, average ± SD; Figure 5A). Of all bone marrow CD79α+ B lymphocytes, 11±2.5% expressed TdT. In contrast, in the fetal lymph node, 29±18% of the TdT positive cells were B lymphocytes, 33±14% were T lymphocytes, and only 0.14±0.08% of the B lymphocytes expressed TdT (n = 4; Figure 5B). Fetal spleen, liver and ileal Peyer's patch, as well as adult lymph node and spleen contained very few TdT+ B cells (0.002–0.04% of all CD79α+ B cells). B lymphocytes were rare in adult bone marrow and liver.
Figure 5. Immunofluorescence staining of fetal bovine bone marrow (A) and lymph node (B).
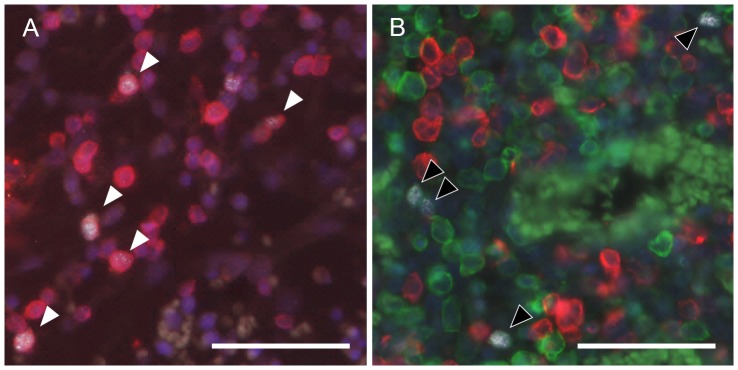
Red: B lymphocyte marker CD79α. Green: T lymphocyte marker CD3. White: TdT. Blue: DAPI. White arrows: TdT positive B cells. Black arrows: TdT positive T cells. Scale bar: 50 µm.
Discussion
To complement their restricted range of immunoglobulin genes [29], [30], [38], [39], fetal cattle use two main mechanisms to secure a functional preimmune antibody repertoire: AID-driven somatic hypermutation [6] and TdT-mediated junctional diversity that is the focus of this paper. We first searched for potentially new immunoglobulin genes, as assessing junctional diversity is dependent on the accurate definition of V, D, and J gene segment boundaries in the rearranged immunoglobulin genes. We then analyzed the junctions between V, D, and J segments. Finally, we characterized the expression of TdT and its isoforms in fetal tissues.
Bovine immunoglobulin heavy chain variable and diversity genes
We complied with the IMGT recommendations [37] for naming the immunoglobulin genes (Tables S1 and S2). IGHV1S37 has been previously published [40] with accession JN897034, and several of the other sequences differed from the previously published ones by one nucleotide only. To ease the comparisons with the previously identified IGHD genes, all the currently known IGHD genes were summarized (Table S1) and the old gene names (DH1 to DH5, D64, DH7, DH8, and DQ52) [7], [28] were retained next to those complying with IMGT recommendations (IGHDS1 to IGHDS9 and IGHDS10 to IGHDS14 corresponding to the previously known and new IGHD genes, respectively).
Twenty-six new potentially functional IGHV genes representing a single subgroup were identified from muscle genomic DNA (Table S1). Together with the previously reported 10 functional genes [36], the total number of potentially functional IGHV genes in our material was 36 of which a maximum of 20 were found from a single animal. However, these gene sequences represent both actual paralogous genes and allelic variants, which cannot be distinguished only based on gene sequence data [41]. As there is a maximum of two alleles per locus in a diploid genome, these observations suggest that cattle have 10 to 20 functional paralogous IGHV genes in total (presuming 100% heterozygosity or 100% homozygosity, respectively).
Long CDR3Hs and long IGHDs are well documented in cattle immunoglobulins [3], [5], [7], [42]. In our data from fetal bone marrow, ileum and spleen, the long IGHD genes (>100 nucleotides) were utilized in 13% of the recombinations. It was recently proposed that the domains of “ultra long” CDR3Hs form a unique antibody structure. The authors also described the longest CDR3H to date with 67 codons [43]. The CDR3H encoding regions in our material ranged from 5 to 65 codons. The longest CDR3Hs were encoded by IGHV1S1 or IGHV1S15, IGHDS12 or IGHDS2 and JH1. IGHV1S1 and IGHV1S15 code for an unusual “TTVHQ” terminal motif, which initiates an ascending β strand in the folded antibody [43]. To date, these long CDR3H populations have not been detected in sheep or swine [44], [45].
To uncover new bovine IGHD genes we searched the entire high throughput genomic and trace archive databases at NCBI using RSS motives as queries. The utilized RSS motives identify all published bovine RSS sequences and also most of the published human RSSs. The novel IGHDS14 gene uncovered from the sequenced cDNAs could not be found in the archives. It is possible that additional IGHD genes or allelic variants are present in the archives and thus remain to be discovered. The correct assignment of a particular D segment to cDNAs with a short CDR3H by best pairwise alignment score is further compromised by the short sequence motifs shared by several D segments (Table S2).
Size variation in bovine immunoglobulin heavy and light chain N regions
We detected additions of several N nucleotides in most fetal IGH cDNA sequences suggesting that TdT is active in fetal life. Exceptionally long (10–36 bp) N nucleotide additions were detected in 4.0% of VD and 3.2% of DJ junctions. Such long N additions have not been reported in other species to our knowledge. In contrast, N regions longer than 10 nucleotides are considered abnormal in mice [46], [47]. Nontemplated additions to IGH genes have been reported in fetal humans [48], swine [44], [49] and sheep [45] but not in fetal or neonatal mouse [50]. In humans 1-6.9 N nucleotides are added on average, depending on the IGHD gene used [48]. Also, N additions in fetal swine are fairly common. Average number is 7.9–9.9 nucleotides and can reach up to 20 nucleotides per coding joint [44], [49]. In sheep, the exact analysis of the junctional variability has not been possible because of the lack of knowledge of the IGHD genes [45]. Despite the great range of N additions in our data, their average number in cattle was not especially high (2.5/VD junction and 2.6/DJ junction) compared to other species, reflecting the high frequency (35%) of junctions with zero additions.
Nontemplated nucleotide additions were also present in IGL and IGK light chain genes although their number was lower than in heavy chain genes. Junctional diversity has been observed in the ovine IGK light chains. In ovine IGL genes, the extent of junctional diversity appears to depend on the specific joining gene used [51], [52]. Very little junctional diversity is seen in swine light chains [53], [54]. In human and mouse junctional diversity is absent in light chains, as TdT expression is restricted to the pro-B stages. The expression starts to weaken already during pre-B cell stage and is absent in later stages when light chain rearrangements occur [55], [56].
In addition to N-nucleotide additions, the junctional regions were modified by exonuclease activity targeted to IGL and IGH gene ends. The median number of removed nucleotides ranged from 1 to 3 for the variable and joining gene ends. In contrast, the trimming of the IGHD gene ends appeared more extensive (median value of 5 and 6 nucleotides for the VD and JD junction, respectively). Extensive trimming of IGHD genes is also observed in swine where the longer porcine DHA was trimmed to the same length as the shorter DHB [44]. Protein conformation tolerates the trimming of IGHD better than that of IGHV or IGHJ, because IGHD does not encode for framework regions. In our data, there were 63 (10%) cDNAs where the number of removed nucleotides from either junction was greater than 29. Removal of these cDNAs did not affect the median or range of P or N nucleotide additions presented in Table 2. However, IGHDS2 was assigned to 31 of these cDNAs (Table 6). The frequency of IGHDS2 in recombinations (Table 2) has to be interpreted with great caution.
Table 6. The effect of removal of 63 cDNAs linked to high exonuclease activity to the frequency of IGHD segments in bovine fetal immunoglobulin cDNAs.
| IGHD | High exo | Normal exo | Sum |
| IGHDS1 | 1 | 3 | 4 |
| IGHDS2 | 31 | 10 | 41 |
| IGHDS3 | 7 | 85 | 92 |
| IGHDS4 | 0 | 24 | 24 |
| IGHDS5 | 14 | 254 | 268 |
| IGHDS6 | 0 | 1 | 1 |
| IGHDS7 | 0 | 79 | 79 |
| IGHDS8 | 0 | 71 | 71 |
| IGHDS9 | 0 | 9 | 9 |
| IGHDS10 | 0 | 3 | 3 |
| IGHDS11 | 5 | 4 | 9 |
| IGHDS12 | 2 | 7 | 9 |
| IGHDS13 | 0 | 3 | 3 |
| IGHDS14 | 3 | 29 | 32 |
| Sum | 63 | 582 | 645 |
Exonuclease activity was deduced from the alignments with the best matching IGHD segment and quantified by the number of apparently excised nucleotides. High exonuclease activity: excision of over 29 nucleotides from either end of the IGHD segment.
We defined the D-region boundaries on the basis of the coordinates of the best pairwise alignment between the cDNA sequence and IGHD genes. To ensure that only nontemplated and P nucleotides are included in the VD and DJ joints, the extension of gaps was only marginally penalized. The presence of gaps in the alignments may indicate that the existence of additional IGHD genes or alleles. This will not affect the overall conclusions of this work since the total number of sequences with gaps in the alignment was 11 (2%). Alternatively, SHM process might induce small insertions and deletions. Also, IGHD genes contain repetitive TAT and GGT codons that could be incorrectly copied by the cellular DNA polymerases either during SHM or DNA replication.
TdT expression in fetal cattle
We analysed the fetal expression of TdT while adult thymus served as a positive control [57], [58]. Of extra-thymic tissues, bone marrow displayed the strongest TdT expression with more than 10% of all B cells being TdT positive. TdT expression was also detected in fetal lymph nodes, but this was largely due to TdT positive T cells; only a very small fraction of lymph node B cells expressed TdT (Figures 4 and 5). A significant number of TdT positive cells were negative for both CD markers. These possibly represent lymphoid progenitor cells, which sometimes express TdT already at the CD34+ stage [59]. In fact, TdT was originally considered a marker for immature lymphoid cells [60]. TdT expression has also been shown to be associated with acute myeloid leukemia, suggesting that TdT expression is not always limited to cells fully committed to the lymphoid lineage [61], [62]. This may also explain our finding that TdT expression, as measured by qPCR, is at a similar level in the adult and fetal bone marrow as shown in Figure 4. In adults, B cells are very rare and de novo B lymphopoiesis has practically ceased [23].
Alternative splicing occurs in TdT. Long splice variants (TdTLs), which possess an extra exon forming a new catalytic site for the enzyme, have been suggested to have exonuclease activity in humans [21]. Like humans, cattle have three potential isoforms. In addition to the shorter form (bTdTS), two longer fragments (bTdTL1 and bTdTL2) have been found from bovine thymic cDNA [20]. We detected the long variants mainly in thymus while the expression levels in other tissues were low. This suggests that long isoforms could be mainly T-cell specific in cattle.
TdT and N-nucleotide additions in fetal cattle
TdT is sufficient for N-region diversity in mouse immunoglobulin loci [46]. However, the very long N additions observed here differ from the murine TdT signature [46], [47]. Also, the bias towards T additions is in contrast to the previous findings of 60–70% of dGMP residues in N additions in vitro [13]. There are plausible explanations for these differences. First, the G/C nucleotide bias is less emphasized in long extensions. The dGMP nucleotides tend to form aggregates resulting in the 3′-OH group of the growing polymer becoming relatively less accessible to further chain growth [63], [64]. This suggests that long G/C rich additions may be disfavored in vivo due to conformational restrictions. Second, the function of TdT is known to be dissimilar in vivo versus in vitro. Mouse in vivo studies show 2–5 nucleotide additions [65] while in vitro TdT can add several kilobases of nucleotides under optimal conditions [12]. DNA-PK limits the length of TdT-induced nucleotide additions in vitro by reducing the number of modified DNA ends and the length of nucleotide additions [64]. More recently, Ku80, which is a part of DNA-PK, was shown to inhibit the DNA strand elongation activity by TdT [47]. DNA-PK or Ku80 proteins have not been investigated in cattle so it remains to be resolved whether or not these components are involved in regulating the bovine TdT activity. The lack a TdT inhibitor would give better access to the free coding ends during V(D)J recombination, promote more efficient initiation of polymerization and lead to a greater number of modified V(D)J ends. Alternatively, it could permit longer nucleotide additions than seen in human or mouse by increasing the processivity of TdT.
In addition to TdT, other polymerases of the PolX family may also contribute to junctional diversity. Polµ deficient mice have about 6 bp shorter VJ junctions in κ light chains compared to wild type, suggesting that Polµ takes part in immunoglobulin gene diversification after TdT expression has decreased [66]. In mice Polµ is active during early embryonic DJH rearrangements. It can perform template-independent nucleotide additions in a similar manner to TdT [16]. Also Polλ polymerase functions in heavy chain rearrangements [67]. Apart from TdT, other PolX family members are currently uncharacterized in cattle and require further investigation.
In conclusion, our data suggest that junctional diversity plays a significant role in the generation of the bovine preimmune immunoglobulin repertoire. TdT is expressed in fetal bone marrow B cells in conjunction with the recombination machinery [22], [23]. The analysis of immunoglobulin cDNA sequences confirms the diversification of the V(D)J junctions by a combination of polymerase and exonuclease activities. According to the prevailing model of B-cell development in ruminants [2], a subpopulation of B cells expressing the B cell receptor then seeds the ileal Peyer's patch where somatic hypermutation further diversifies the repertoire. Together, junctional diversity and somatic hypermutation complement the small range of immunoglobulin genes and enable the creation of a sufficiently large functional preimmune repertoire during late fetal life.
Supporting Information
Alignment views of immunoglobulin heavy chain sequences with IGHD gene segments. Initial alignments of 645 immunoglobulin cDNAs to the best matching reference gene from IGHDS1 to IGHDS13. Boundaries of the CDR3H region as defined by IMGT and putative amino acid sequence are indicated. Note that many of the cDNAs matching to IGHDS12 contain a novel gene segment IGHDS14 (see also Figure S2).
(TXT)
Deduction of novel IGHDS14 sequence. A set of cDNA sequences was selected based on pairwise alignments with IGHDS12 in Figure S1. Regions between V and J segments were extracted and aligned with MUSCLE [26]. Consensus sequence corresponding to IGHDS14 is shown. Star (*) indicates a completely conserved nucleotide.
(TXT)
New bovine IGHV sequences characterized in this study. The sequences have been submitted to GenBank (accessions KJ491073-KJ491098). IGHVS18-IGHVS40 contain first eight bases of RSS (typed in lower case).
(DOCX)
General information on bovine gene segments IGHDS1-IGHDS14.
(XLSX)
General information on bovine gene segments JH1, JH2 and JH6.
(XLSX)
Sequence analysis of CDR3H region. Nucleotides corresponding to V, D and J gene segments, N and P nucleotides in VD and DJ junctions, and putative peptide sequence are shown for 645 fetal bovine immunoglobulin cDNAs. Small letters: framework nucleotides. Capital letters: nucleotides corresponding to CDR3H region.
(XLSX)
Quantification of N and P nucleotide additions and exonuclease activity during immunoglobulin recombination at the heavy chain locus. The VDJ junctions of 645 bovine fetal immunoglobulin cDNAs were analyzed from pairwise alignments with best matching reference V, D, and J gene segments. The number of added (N and P) nucleotides and the number of removed nucleotides from each reference gene end are shown.
(XLSX)
Quantification of N and P nucleotide additions and exonuclease activity during immunoglobulin recombination and the λ light chain locus. The VJ junctions of 65 bovine fetal λ light chain cDNAs were analyzed from pairwise alignments with best matching reference V and J gene segments. The number of added (N and P) nucleotides and the number of removed nucleotides from each reference gene end are shown.
(XLSX)
Quantification of N and P nucleotide additions and exonuclease activity during immunoglobulin recombination at the κ light chain locus. The VJ junctions of 83 bovine fetal κ light chain cDNAs were analyzed from pairwise alignments with best matching reference V and J gene segments. The number of added (N and P) nucleotides and the number of removed nucleotides from each reference gene end are shown.
(XLSX)
Acknowledgments
We thank Kirsi Lahti and Tuire Pankasalo for their excellent technical assistance, Else Anttila for helping to collect the fetal material, and Robert Leigh for comments on the manuscript.
Funding Statement
This study was supported by The Academy of Finland, Finnish Veterinary Foundation, Finnish Foundation of Veterinary Research, and Orion-Farmos Research Foundation. JL is a student at the Viikki Graduate School in Molecular Biosciences. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Tonegawa S (1983) Somatic generation of antibody diversity. Nature 302: 575–581. [DOI] [PubMed] [Google Scholar]
- 2. Weill JC, Reynaud CA (1998) Galt versus bone marrow models of B cell ontogeny. Dev Comp Immunol 22: 379–385. [DOI] [PubMed] [Google Scholar]
- 3. Saini SS, Allore B, Jacobs RM, Kaushik A (1999) Exceptionally long CDR3H region with multiple cysteine residues in functional bovine IgM antibodies. Eur J Immunol 29: 2420–2426. [DOI] [PubMed] [Google Scholar]
- 4. Koti M, Kataeva G, Kaushik AK (2008) Organization of D(H)-gene locus is distinct in cattle. Dev Biol (Basel) 132: 307–313. [DOI] [PubMed] [Google Scholar]
- 5. Kaushik AK, Kehrli ME Jr, Kurtz A, Ng S, Koti M, et al. (2009) Somatic hypermutations and isotype restricted exceptionally long CDR3H contribute to antibody diversification in cattle. Vet Immunol Immunopathol 127: 106–113 10.1016/j.vetimm.2008.09.024 [DOI] [PubMed] [Google Scholar]
- 6.Liljavirta J, Ekman A, Knight JS, Pernthaner A, Iivanainen A, et al.. (2013) Activation-induced cytidine deaminase (AID) is strongly expressed in the fetal bovine ileal Peyer's patch and spleen and is associated with expansion of the primary antibody repertoire in the absence of exogenous antigens. Mucosal Immunol. doi:10.1038/mi.2012.132. [DOI] [PubMed]
- 7. Koti M, Kataeva G, Kaushik AK (2010) Novel atypical nucleotide insertions specifically at VH-DH junction generate exceptionally long CDR3H in cattle antibodies. MolImmunol 47: 2119–2128 10.1016/j.molimm.2010.02.014 [DOI] [PubMed] [Google Scholar]
- 8. Gauss GH, Lieber MR (1996) Mechanistic constraints on diversity in human V(D)J recombination. Mol Cell Biol 16: 258–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murphy KM (2012) Janeway's Immunobiology, 8th Edition (Immunobiology: The Immune System. 8th ed. Garland Science. 888 p. [Google Scholar]
- 10. Alt FW, Baltimore D (1982) Joining of immunoglobulin heavy chain gene segments: implications from a chromosome with evidence of three D-JH fusions. Proc Natl Acad Sci USA 79: 4118–4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Komori T, Okada A, Stewart V, Alt FW (1993) Lack of N regions in antigen receptor variable region genes of TdT-deficient lymphocytes. Science 261: 1171–1175. [DOI] [PubMed] [Google Scholar]
- 12. Chang LM, Bollum FJ (1986) Molecular biology of terminal transferase. CRC Crit Rev Biochem 21: 27–52. [DOI] [PubMed] [Google Scholar]
- 13. Basu M, Hegde MV, Modak MJ (1983) Synthesis of compositionally unique DNA by terminal deoxynucleotidyl transferase. Biochem Biophys Res Commun 111: 1105–1112. [DOI] [PubMed] [Google Scholar]
- 14. Greenberg JM, Kersey JH (1987) Terminal deoxynucleotidyl transferase expression can precede T cell receptor beta chain and gamma chain rearrangement in T cell acute lymphoblastic leukemia. Blood 69: 356–360. [PubMed] [Google Scholar]
- 15. Uchiyama Y, Takeuchi R, Kodera H, Sakaguchi K (2009) Distribution and roles of X-family DNA polymerases in eukaryotes. Biochimie 91: 165–170 10.1016/j.biochi.2008.07.005 [DOI] [PubMed] [Google Scholar]
- 16. Gozalbo-López B, Andrade P, Terrados G, de Andrés B, Serrano N, et al. (2009) A role for DNA polymerase mu in the emerging DJH rearrangements of the postgastrulation mouse embryo. Mol Cell Biol 29: 1266–1275 10.1128/MCB.01518-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bentolila LA, Fanton d'Andon M, Nguyen QT, Martinez O, Rougeon F, et al. (1995) The two isoforms of mouse terminal deoxynucleotidyl transferase differ in both the ability to add N regions and subcellular localization. EMBO J 14: 4221–4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Benedict CL, Gilfillan S, Thai T-H, Kearney JF (2000) Terminal deoxynucleotidyl transferase and repertoire development. Immunological Reviews 175: 150–157. [PubMed] [Google Scholar]
- 19. Benedict CL, Gilfillan S, Kearney JF (2001) The long isoform of terminal deoxynucleotidyl transferase enters the nucleus and, rather than catalyzing nontemplated nucleotide addition, modulates the catalytic activity of the short isoform. J Exp Med 193: 89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Takahara K, Hayashi N, Fujita-Sagawa K, Morishita T, Hashimoto Y, et al. (1994) Alternative splicing of bovine terminal deoxynucleotidyl transferase cDNA. Biosci Biotechnol Biochem 58: 786–787. [DOI] [PubMed] [Google Scholar]
- 21. Thai T-H, Kearney JF (2004) Distinct and opposite activities of human terminal deoxynucleotidyltransferase splice variants. J Immunol 173: 4009–4019. [DOI] [PubMed] [Google Scholar]
- 22. Ekman A, Pessa-Morikawa T, Liljavirta J, Niku M, Iivanainen A (2010) B-cell development in bovine fetuses proceeds via a pre-B like cell in bone marrow and lymph nodes. Dev Comp Immunol 34: 896–903 10.1016/j.dci.2010.03.012 [DOI] [PubMed] [Google Scholar]
- 23. Ekman A, Ilves M, Iivanainen A (2012) B lymphopoiesis is characterized by pre-B cell marker gene expression in fetal cattle and declines in adults. Dev Comp Immunol 37: 39–49 10.1016/j.dci.2011.12.009 [DOI] [PubMed] [Google Scholar]
- 24. Rice P, Longden I, Bleasby A (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16: 276–277. [DOI] [PubMed] [Google Scholar]
- 25.R Development Core Team (2013) R: A Language and Environment for Statistical Computing. Vienna, Austria. Available: http://www.R-project.org/.
- 26. Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhao Y, Kacskovics I, Rabbani H, Hammarstrom L (2003) Physical mapping of the bovine immunoglobulin heavy chain constant region gene locus. J Biol Chem 278: 35024–35032 10.1074/jbc.M301337200 [DOI] [PubMed] [Google Scholar]
- 28. Hosseini A, Campbell G, Prorocic M, Aitken R (2004) Duplicated copies of the bovine JH locus contribute to the Ig repertoire. Int Immunol 16: 843–852 10.1093/intimm/dxh085 [DOI] [PubMed] [Google Scholar]
- 29. Berens SJ, Wylie DE, Lopez OJ (1997) Use of a single VH family and long CDR3s in the variable region of cattle Ig heavy chains. Int Immunol 9: 189–199. [DOI] [PubMed] [Google Scholar]
- 30. Ekman A, Niku M, Liljavirta J, Iivanainen A (2009) Bos taurus genome sequence reveals the assortment of immunoglobulin and surrogate light chain genes in domestic cattle. BMC Immunol 10: 22 10.1186/1471-2172-10-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pages H, Aboyoun P, Gentleman R, DebRoy S (2014) Biostrings: String objects representing biological sequences, and matching algorithms. R package version 2.30.1.
- 32. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 33. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 34.Hollander M, Wolfe DA (1999) Nonparametric Statistical Methods. 2nd ed. New York, NY, USA: John Wiley & Sons, Ltd. 816 p. [Google Scholar]
- 35. Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Niku M, Liljavirta J, Durkin K, Schroderus E, Iivanainen A (2012) The bovine genomic DNA sequence data reveal three IGHV subgroups, only one of which is functionally expressed. Developmental & Comparative Immunology 37: 457–461 10.1016/j.dci.2012.02.006 [DOI] [PubMed] [Google Scholar]
- 37.Lefranc MP (2001) Nomenclature of the human immunoglobulin genes. Curr Protoc Immunol Appendix 1: Appendix 1P. doi:10.1002/0471142735.ima01ps40. [DOI] [PubMed]
- 38. Saini SS, Hein WR, Kaushik A (1997) A single predominantly expressed polymorphic immunoglobulin V(H) gene family, related to mammalian group, I, clan, II, is identified in cattle. MolImmunol 34: 641–651 10.1016/S0161-5890(97)00055-2 [DOI] [PubMed] [Google Scholar]
- 39. Sinclair MC, Gilchrist J, Aitken R (1997) Bovine IgG repertoire is dominated by a single diversified VH gene family. J Immunol 159: 3883–3889. [PubMed] [Google Scholar]
- 40. Verma S, Aitken R (2012) Somatic hypermutation leads to diversification of the heavy chain immunoglobulin repertoire in cattle. Vet Immunol Immunopathol 145: 14–22 10.1016/j.vetimm.2011.10.001 [DOI] [PubMed] [Google Scholar]
- 41. Pramanik S, Cui X, Wang H-Y, Chimge N-O, Hu G, et al. (2011) Segmental duplication as one of the driving forces underlying the diversity of the human immunoglobulin heavy chain variable gene region. BMC Genomics 12: 78 10.1186/1471-2164-12-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhao Y, Jackson SM, Aitken R (2006) The bovine antibody repertoire. Dev Comp Immunol 30: 175–186 10.1016/j.dci.2005.06.012 [DOI] [PubMed] [Google Scholar]
- 43. Wang F, Ekiert DC, Ahmad I, Yu W, Zhang Y, et al. (2013) Reshaping antibody diversity. Cell 153: 1379–1393 10.1016/j.cell.2013.04.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Butler JE, Weber P, Sinkora M, Sun J, Ford SJ, et al. (2000) Antibody repertoire development in fetal and neonatal piglets. II. Characterization of heavy chain complementarity-determining region 3 diversity in the developing fetus. J Immunol 165: 6999–7010. [DOI] [PubMed] [Google Scholar]
- 45. Gontier E, Ayrault O, Godet I, Nau F, Ladevèze V (2005) Developmental progression of immunoglobulin heavy chain diversity in sheep. Vet Immunol Immunopathol 103: 31–51 10.1016/j.vetimm.2004.08.013 [DOI] [PubMed] [Google Scholar]
- 46. Bentolila LA, Wu GE, Nourrit F, Fanton d'Andon M, Rougeon F, et al. (1997) Constitutive expression of terminal deoxynucleotidyl transferase in transgenic mice is sufficient for N region diversity to occur at any Ig locus throughout B cell differentiation. J Immunol 158: 715–723. [PubMed] [Google Scholar]
- 47. Sandor Z, Calicchio ML, Sargent RG, Roth DB, Wilson JH (2004) Distinct requirements for Ku in N nucleotide addition at V(D)J- and non-V(D)J-generated double-strand breaks. Nucleic Acids Res 32: 1866–1873 10.1093/nar/gkh502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schroeder HW Jr, Mortari F, Shiokawa S, Kirkham PM, Elgavish RA, et al. (1995) Developmental regulation of the human antibody repertoire. Ann N Y Acad Sci 764: 242–260. [DOI] [PubMed] [Google Scholar]
- 49. Sinkora M, Sun J, Sinkorová J, Christenson RK, Ford SP, et al. (2003) Antibody repertoire development in fetal and neonatal piglets. VI. B cell lymphogenesis occurs at multiple sites with differences in the frequency of in-frame rearrangements. J Immunol 170: 1781–1788. [DOI] [PubMed] [Google Scholar]
- 50. Feeney AJ (1990) Lack of N regions in fetal and neonatal mouse immunoglobulin V-D-J junctional sequences. J Exp Med 172: 1377–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jeong Y, Osborne BA, Goldsby RA (2001) Early Vlambda diversification in sheep. Immunology 103: 26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jenne CN, Kennedy LJ, McCullagh P, Reynolds JD (2003) A new model of sheep Ig diversification: shifting the emphasis toward combinatorial mechanisms and away from hypermutation. J Immunol 170: 3739–3750. [DOI] [PubMed] [Google Scholar]
- 53. Butler JE, Wertz N, Sun J, Wang H, Chardon P, et al. (2004) Antibody repertoire development in fetal and neonatal pigs. VII. Characterization of the preimmune kappa light chain repertoire. J Immunol 173: 6794–6805. [DOI] [PubMed] [Google Scholar]
- 54. Wertz N, Vazquez J, Wells K, Sun J, Butler JE (2013) Antibody repertoire development in fetal and neonatal piglets. XII. Three IGLV genes comprise 70% of the pre-immune repertoire and there is little junctional diversity. Mol Immunol 55: 319–328 10.1016/j.molimm.2013.03.012 [DOI] [PubMed] [Google Scholar]
- 55. Galler GR, Mundt C, Parker M, Pelanda R, Mårtensson I-L, et al. (2004) Surface mu heavy chain signals down-regulation of the V(D)J-recombinase machinery in the absence of surrogate light chain components. J Exp Med 199: 1523–1532 10.1084/jem.20031523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li YS, Hayakawa K, Hardy RR (1993) The regulated expression of B lineage associated genes during B cell differentiation in bone marrow and fetal liver. J Exp Med 178: 951–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gregoire KE, Goldschneider I, Barton RW, Bollum FJ (1979) Ontogeny of terminal deoxynucleotidyl transferase-positive cells in lymphohemopoietic tissues of rat and mouse. J Immunol 123: 1347–1352. [PubMed] [Google Scholar]
- 58. Deibel MR Jr, Riley LK, Coleman MS, Cibull ML, Fuller SA, et al. (1983) Expression of terminal deoxynucleotidyl transferase in human thymus during ontogeny and development. J Immunol 131: 195–200. [PubMed] [Google Scholar]
- 59. Gore SD, Kastan MB, Civin CI (1991) Normal human bone marrow precursors that express terminal deoxynucleotidyl transferase include T-cell precursors and possible lymphoid stem cells. Blood 77: 1681–1690. [PubMed] [Google Scholar]
- 60. Desiderio SV, Yancopoulos GD, Paskind M, Thomas E, Boss MA, et al. (1984) Insertion of N regions into heavy-chain genes is correlated with expression of terminal deoxytransferase in B cells. Nature 311: 752–755. [DOI] [PubMed] [Google Scholar]
- 61. Drexler HG, Sperling C, Ludwig WD (1993) Terminal deoxynucleotidyl transferase (TdT) expression in acute myeloid leukemia. Leukemia 7: 1142–1150. [PubMed] [Google Scholar]
- 62. Patel KP, Khokhar FA, Muzzafar T, James You M, Bueso-Ramos CE, et al. (2013) TdT expression in acute myeloid leukemia with minimal differentiation is associated with distinctive clinicopathological features and better overall survival following stem cell transplantation. Mod Pathol 26: 195–203 10.1038/modpathol.2012.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lefler CF, Bollum FJ (1969) Deoxynucleotide-polymerizing enzymes of calf thymus gland. 3. Preparation of poly N-acetyldeoxyguanylate and polydeoxyguanylate. J Biol Chem 244: 594–601. [PubMed] [Google Scholar]
- 64. Mickelsen S, Snyder C, Trujillo K, Bogue M, Roth DB, et al. (1999) Modulation of terminal deoxynucleotidyltransferase activity by the DNA-dependent protein kinase. J Immunol 163: 834–843. [PubMed] [Google Scholar]
- 65. Gilfillan S, Benoist C, Mathis D (1995) Mice lacking terminal deoxynucleotidyl transferase: adult mice with a fetal antigen receptor repertoire. Immunol Rev 148: 201–219. [DOI] [PubMed] [Google Scholar]
- 66. Bertocci B, De Smet A, Berek C, Weill J-C, Reynaud C-A (2003) Immunoglobulin kappa light chain gene rearrangement is impaired in mice deficient for DNA polymerase mu. Immunity 19: 203–211. [DOI] [PubMed] [Google Scholar]
- 67. Bertocci B, De Smet A, Weill J-C, Reynaud C-A (2006) Nonoverlapping functions of DNA polymerases mu, lambda, and terminal deoxynucleotidyltransferase during immunoglobulin V(D)J recombination in vivo. Immunity 25: 31–41 10.1016/j.immuni.2006.04.013 [DOI] [PubMed] [Google Scholar]
- 68. Riley LK, Morrow JK, Danton MJ, Coleman MS (1988) Human terminal deoxyribonucleotidyltransferase: molecular cloning and structural analysis of the gene and 5′ flanking region. Proc Natl Acad Sci USA 85: 2489–2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment views of immunoglobulin heavy chain sequences with IGHD gene segments. Initial alignments of 645 immunoglobulin cDNAs to the best matching reference gene from IGHDS1 to IGHDS13. Boundaries of the CDR3H region as defined by IMGT and putative amino acid sequence are indicated. Note that many of the cDNAs matching to IGHDS12 contain a novel gene segment IGHDS14 (see also Figure S2).
(TXT)
Deduction of novel IGHDS14 sequence. A set of cDNA sequences was selected based on pairwise alignments with IGHDS12 in Figure S1. Regions between V and J segments were extracted and aligned with MUSCLE [26]. Consensus sequence corresponding to IGHDS14 is shown. Star (*) indicates a completely conserved nucleotide.
(TXT)
New bovine IGHV sequences characterized in this study. The sequences have been submitted to GenBank (accessions KJ491073-KJ491098). IGHVS18-IGHVS40 contain first eight bases of RSS (typed in lower case).
(DOCX)
General information on bovine gene segments IGHDS1-IGHDS14.
(XLSX)
General information on bovine gene segments JH1, JH2 and JH6.
(XLSX)
Sequence analysis of CDR3H region. Nucleotides corresponding to V, D and J gene segments, N and P nucleotides in VD and DJ junctions, and putative peptide sequence are shown for 645 fetal bovine immunoglobulin cDNAs. Small letters: framework nucleotides. Capital letters: nucleotides corresponding to CDR3H region.
(XLSX)
Quantification of N and P nucleotide additions and exonuclease activity during immunoglobulin recombination at the heavy chain locus. The VDJ junctions of 645 bovine fetal immunoglobulin cDNAs were analyzed from pairwise alignments with best matching reference V, D, and J gene segments. The number of added (N and P) nucleotides and the number of removed nucleotides from each reference gene end are shown.
(XLSX)
Quantification of N and P nucleotide additions and exonuclease activity during immunoglobulin recombination and the λ light chain locus. The VJ junctions of 65 bovine fetal λ light chain cDNAs were analyzed from pairwise alignments with best matching reference V and J gene segments. The number of added (N and P) nucleotides and the number of removed nucleotides from each reference gene end are shown.
(XLSX)
Quantification of N and P nucleotide additions and exonuclease activity during immunoglobulin recombination at the κ light chain locus. The VJ junctions of 83 bovine fetal κ light chain cDNAs were analyzed from pairwise alignments with best matching reference V and J gene segments. The number of added (N and P) nucleotides and the number of removed nucleotides from each reference gene end are shown.
(XLSX)


