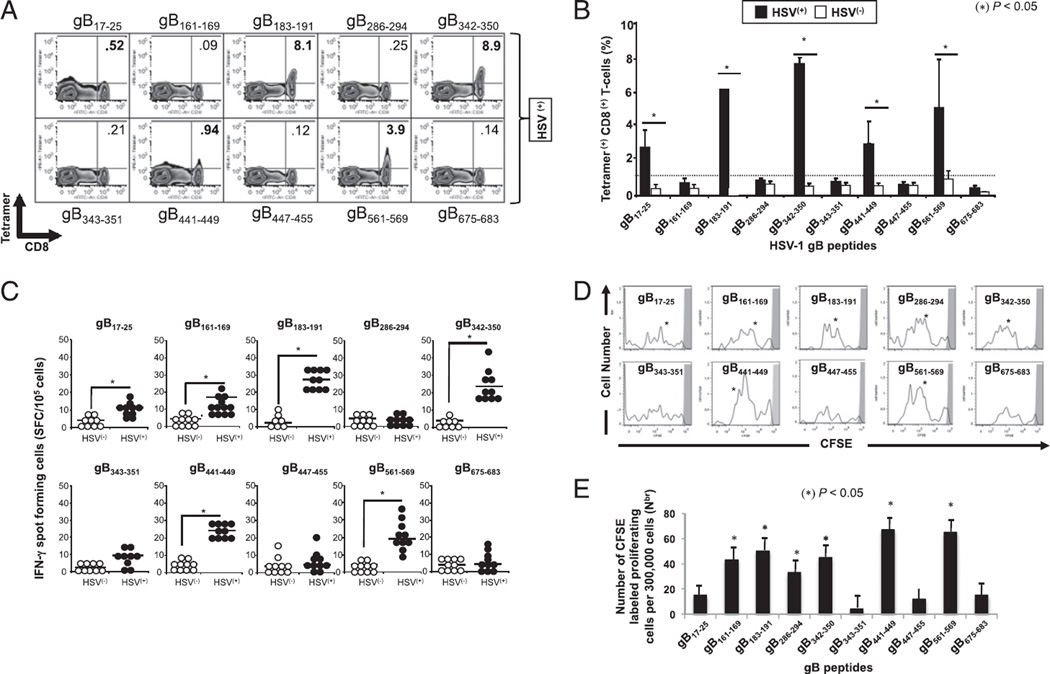
FIGURE 3.
High frequency of IFN-γ–producing CD8+ T cells specific to gB17–25, gB183–191, gB342–350, gB441–449, and gB561–569 epitopes detected in HLA-A*02:01–positive, HSV-seropositive individuals. PBMC-derived CD8+ T cells were analyzed ex vivo, after in vitro expansion, for the frequency of CD8+ T cells specific to 10 gB peptide/tetramer complexes representing each of the 10 potential gB epitopes shown in Fig. 1. (A) Representative FACS data of the frequencies of CD8+ T cells in an HSV-1–seropositive individual following a 5-d expansion with the indicated peptide. (B) Average frequency of PBMC-derived CD8+ T cells that recognize each of the indicated gB epitopes from 10 HLA-A*02:01–positive, HSV-1–seropositive individuals (HSV+) compared with 10 HLA-A*02:01–positive, HSV-seronegative individuals (HSV−). (C) gB epitope-specific IFN-γ–producing CD8+ T cells detected in healthy HLA-A*02:01–positive, HSV-seropositive individuals. PBMC-derived CD8+ T cells, either from HSV+ or from HSV− individuals, were stimulated with individual gB peptides. The number of gB epitope-specific, IFN-γ–producing T cells was determined by ELISPOT assay. Tests were performed in duplicates for each experiment. Spots were developed as described in Materials and Methods and calculated as spot-forming cells = mean number of spots in the presence of Ag − mean number of spots in the absence of stimulation. Open circles represent HSV− individuals whereas filled circles represent HSV+ individuals. (D) Profile of T cell proliferation with individual gB peptides. The graphs represent the proliferation of CD8+ T cells in the presence of 10 µM of each individual gB peptide detected from HSV+ individuals using a CFSE assay. (E) Absolute numbers of dividing CD8+ T cells per 300,000 total cells after 5 d stimulation. The results are representative of two independent experiments. *p < 0.005, comparing HSV+ to HSV− individuals using one-way ANOVA test.