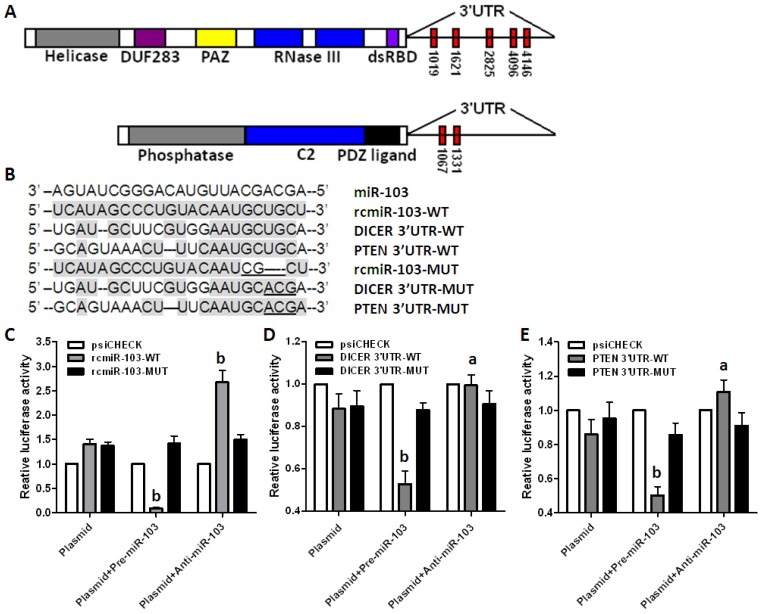
Figure 3.
DICER and PTEN are direct target genes of miR-103. (A) Schematic gene structure of DICER and PTEN and the miR-103 recognition sites located in the 3′-untranslated region (3′-UTR) are shown as red rectangles; (B) Sequence alignment of miR-103 with reverse complementary miR-103 (rcmiR-103-WT), DICER (DICER-3′-UTR-WT), PTEN (PTEN-3′-UTR-WT), mutant rcmiR-103 (rcmiR-103-MUT), mutant DICER (DICER-3′-UTR-MUT), and mutant PTEN (PTEN-3′-UTR-MUT), mutant nucleotides are underlined; and (C–E) Dual-luciferase reporter assay using constructed vectors alone or in the presence of miR-103 precursor or inhibitor was performed. Vectors contain rcmiR-103-WT and rcmiR-103-MUT was used as controls. Renilla luciferase was measured and normalized to Firefly luciferase activity, and the recombinant vector was normalized to empty vector. Data are representative of three experiments. a p < 0.05; b p < 0.01 vs. vector alone group.