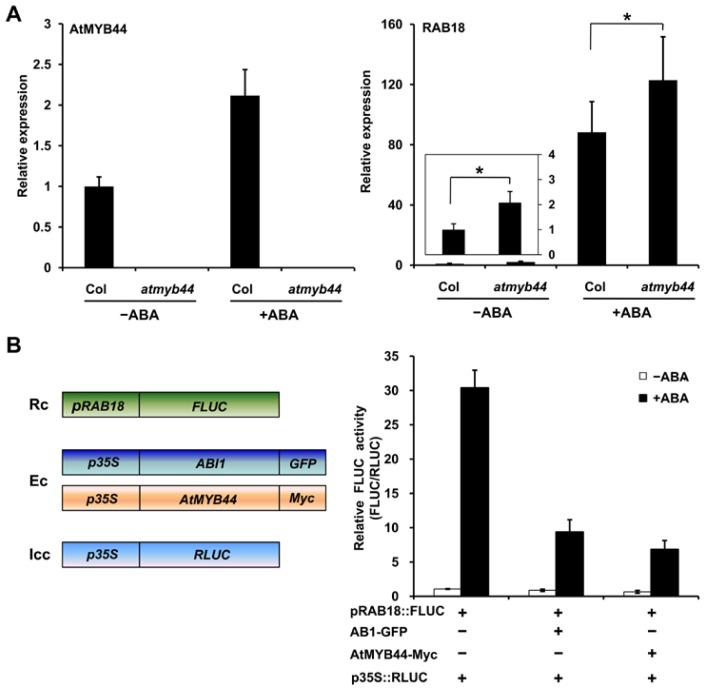
Figure 3.
AtMYB44 negatively regulates the expression of ABA-responsive gene RAB18. (A) Relative expression levels of AtMYB44 and RAB18 in wild-type (Col) and atmyb44 mutant plants were determined by qRT-PCR analysis. 2-week-old seedlings were incubated in 1 × MS liquid medium with ABA (10 μM) or control solvent (DMSO) for 2 h before harvest. ACTIN2/8 served as an internal control. Error bars indicate SD (n = 3). Three independent replicates were analyzed. Asterisks indicate the levels of statistical significance as determined by Student’s t-test: * p < 0.05 vs. Col; and (B) The effect of AtMYB44 on ABA-responsive RAB18 gene expression was analyzed by transactivation assay in Arabidopsis protoplasts. Left panel: Schematic representation of reporter, effector and internal control constructs used in transactivation assays. Rc indicates Reporter construct; Ec indicates Effector constructs; Icc indicates Internal control construct; Right panel: AtMYB44 negatively regulates RAB18 expression in protoplasts. pRAB18::FLUC, p35S::ABI1-GFP, and p35S::AtMYB44-Myc plasmids were co-transfected into Arabidopsis protoplasts from the wild-type plants as the indicated combinations. RLUC was used as an internal control. After transfection, protoplasts were incubated for 5 h under light in the absence (open bars) or presence (filled bars) of 5 μM ABA, and luciferase activity was measured. Values are mean ± SD of three independent experiments.