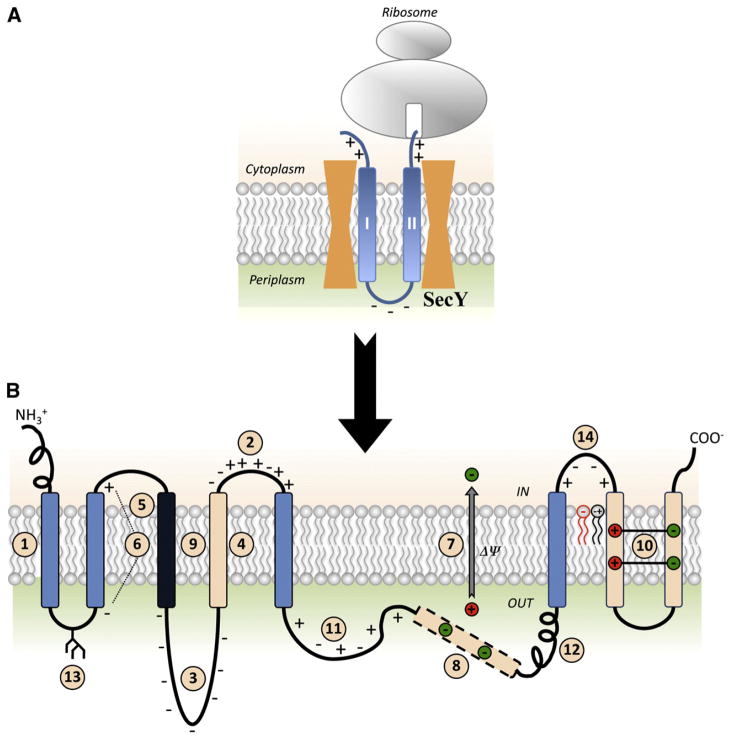
Fig. 1.
Summary of factors determining membrane protein topogenesis and folding. (A) Initial orientation according to the Positive Inside Rule of a nascent two-TMD hairpin is shown in the SecY translocon channel. (B) The features of polytopic membrane proteins and biological membranes that influence folding and final TMD topology. TMDs are color coded for relative hydrophobicity: Light Blue indicates average hydrophobicity; Black indicates above average hydrophobicity; Beige indicates low hydrophobicity. Insertion efficiency of a TMD is driven by overall hydrophobicity with initial orientation determined within the translocon channel (#1) by the Positive Inside Rule (#2) or Charge Difference Rule (#6). Orientation and final topology are influenced by negatively charged residues present in high numbers (#3), flanking a marginally hydrophobic TMD (#4) or that lie at the end of a highly hydrophobic domain (#5) as well as the membrane potential (#7). After exit from the translocon into the lipid bilayer, topology is subject to interactions within the protein where TMDs of low hydrophobicity may become EMDs (#8), highly hydrophobic TMDs stabilize neighboring TMDs of low hydrophobicity (#9), or charged TMDs form salt bridges in the lipid bilayer (#10). Domains with conflicting signals can provide dynamic molecular hinges between independently folding domains (#8 and #11). Rapid, stable folding of an EMD (#12) or co-translational glycosylation of EMDs in eukaryotic cells (#13) can prevent transmembrane shuffling by the translocon. Finally, lipid–protein interactions dictated by the Charge Balance Rule (#14) determine topological organization during initial membrane insertion as well as after folding of membrane proteins.