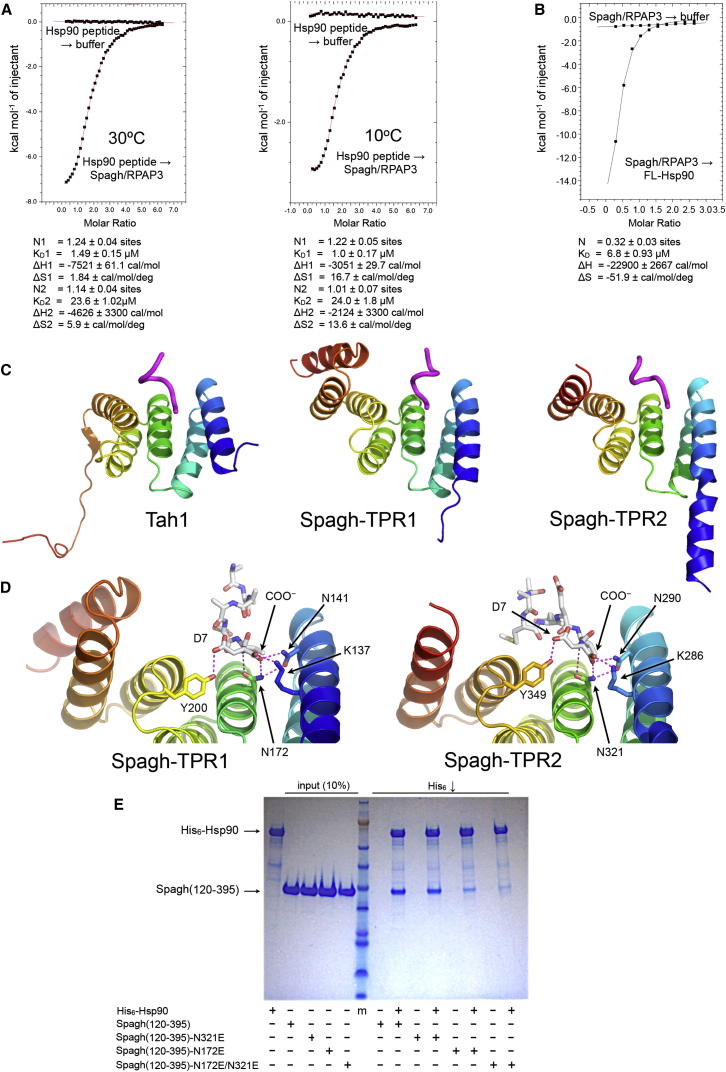
Figure 4.
Structure and Function of Spagh/RPAP3 TPR Domains
(A) ITC binding curve for human Hsp90 C-terminal peptide binding to Spagh/RPAP3 at 30°C (left: KD1 = 1.49 μM and KD2 = 23.6 μM, two site fitting) and 10°C (right: KD1 = 1.0 μM and KD2 = 24 μM, two site fitting). KD estimates are consistent between the two temperatures indicating an accurate fit to the data. The peptide binds in an ∼2:1 (two Hsp90 peptides to one Spagh/RPAP3) molar ratio consistent with Spagh/RPAP3 mutant binding data (Figures S3A and S3B; TPR1, KD = 0.94 μM and TPR2, KD = 9.7 μM).
(B) ITC binding curve for Spagh/RPAP3265–380 binding to full-length Hsp90 dimer with an ∼1:2 molar ratio with KD = 6.8 μM.
(C) Structural comparison of yeast Tah1 TPR-domain (left) and TPR1 (center) and TPR2 domains (right) from Spagh/RPAP3. The N-terminal five helices of the Spagh/RPAP3 TPR domains superimpose with the minimal Tah1-TPR domain. The Cα-trace of the bound Hsp90 C-terminal peptide in each case is shown in magenta.
(D) “Carboxyl” clamp interactions with the C terminus of the Hsp90 tail-peptide by Spagh/RPAP3-TPR1 (left) and TPR2 (right). The C-terminal residue of the peptide is anchored on the side chain of Asn 172 in TPR1 and Asn 321 in TPR2.
(E) Pull-down of Spagh/RPAP3120-395 tandem TPR segment by His6-Hsp90β. Wild-type Spagh/RPAP3 and a TPR1 mutant (N321E) are efficiently coprecipitated, whereas a TPR2 mutant (N172E) diminishes interaction. Mutation of both TPR domains effectively abolishes coprecipitation with Hsp90, confirming the involvement of both TPRs in Hsp90 interaction.