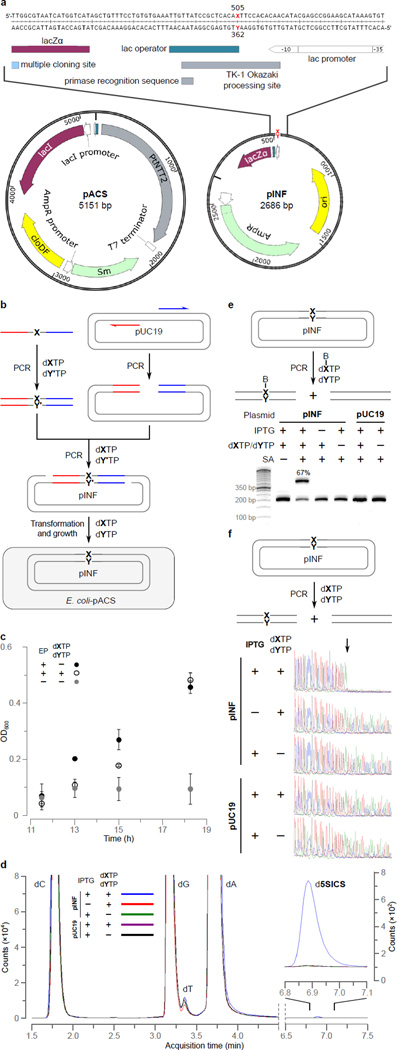
Figure 2. Intracellular UBP replication.
a, Structure of pACS and pINF. dX and dY correspond to dNaM and a d5SICS analog22 that facilitated plasmid construction (see Methods). cloDF = origin of replication; Sm = streptomycin resistance gene; AmpR = ampicillin resistance gene; ori = ColE1 origin of replication; MCS = multiple cloning site; lacZα = β-galactosidase fragment gene. b, Overview of pINF construction. A DNA fragment containing the unnatural nucleotide was synthesized via solid phase DNA synthesis and then used to assemble synthetic pINF via circular-extension PCR29. X = dNaM, Y’ = dTPT3 (an analog of d5SICS22), and Y = d5SICS (see text). Color indicates regions of homology. The doubly-nicked product was used directly to transform E. coli harboring pACS. c, The addition of d5SICSTP and dNaMTP eliminates a growth lag of cells harboring pINF. EP=electroporation. Errors represent s.d. of the mean, n=3. d, LC-MS/MS total ion chromatogram of global nucleoside content in pINF and pUC19 recorded in Dynamic Multiple Reaction Monitoring (DMRM) mode. pINF and pUC19 (control) were propagated in E. coli in the presence or absence of unnatural triphosphates, and with or without PtNTT2 induction. The inset shows a 100× expansion of the mass count axis in the d5SICS region. e, Biotinylation only occurs in the presence of the UBP, the unnatural triphosphates, and transporter induction. After growth, pINF was recovered, and a 194 nt region containing the site of UBP incorporation (nt 437–630) was amplified and biotinylated. B=biotin; SA=streptavidin. The natural pUC19 control plasmid was prepared identically to pINF. 50-bp DNA ladder is shown to the left. f, Sequencing analysis demonstrates retention of the UBP. An abrupt termination in the Sanger sequencing reaction indicates the presence of UBP incorporation (site indicated with arrow).