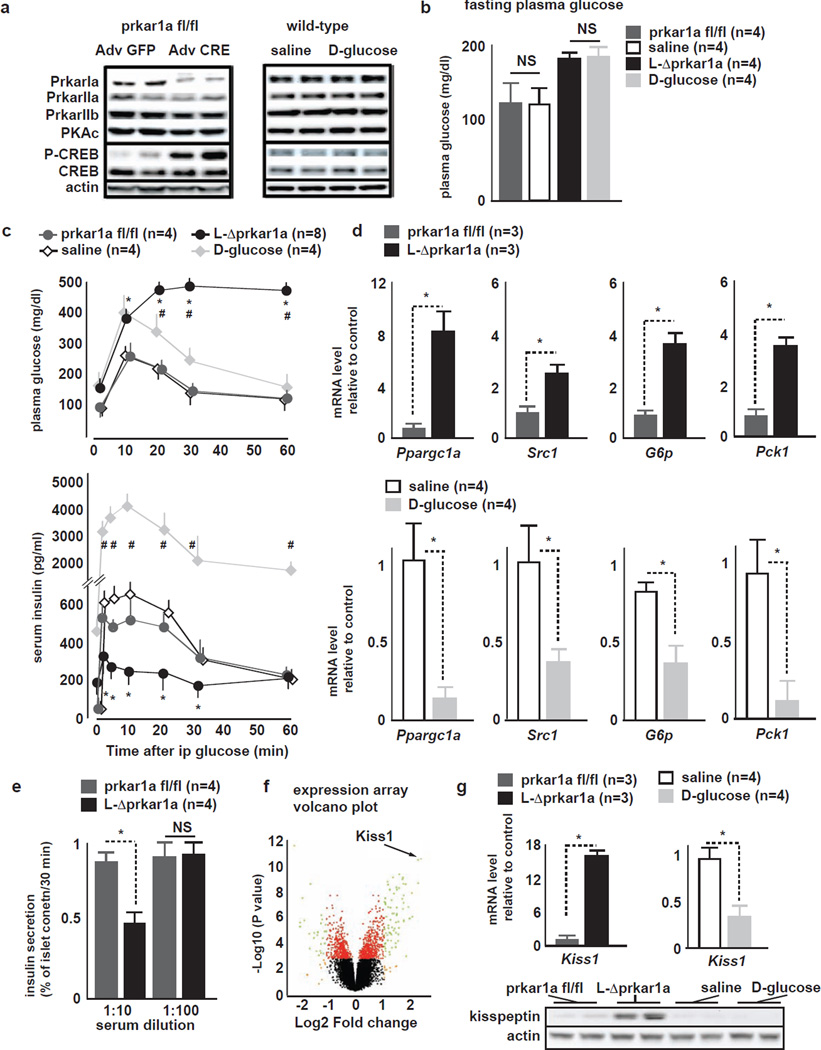
Figure 1. Comparison between L-Δprkar1a and D-glucose mice identifies Kiss1.
A (left) Representative liver IB of prkar1afl/fl and L-Δprkar1a 4 days after adenovirus treatment. L-Δprkar1a mice show Prkar1a ablation and increased pCREB (right) Liver IB from Sal- and D-glucose mice shows unaltered Prkar subtypes, Pkac, pCREB.
B Fasting glucose levels in prkar1afl/fl, L-Δprkar1a, Sal- and D-glucose mice. Prkar1afl/fl and Sal- mice have similar fasting glucose; D-glucose infusion achieves fasting glucose similar to L-Δprkar1a mice (mean±SEM, * P<0.05).
C (top) plasma glucose and (bottom) serum insulin during ipGTT in prkar1afl/fl, L-Δprkar1a, Sal- and D-glucose mice. L-Δprkar1a mice exhibit impaired GT (top) and GSIS (bottom). D-glucose mice have mildly impaired GT and robust GSIS. Prkar1a fl/fl and Sal-mice have similar GT and GSIS (mean±SEM, * P<0.05).
D qRT-PCR of indicated genes of gluconeogenic program in prkar1afl/fl, L-Δprkar1a, Sal- and D-glucose-mouse livers. (top) gluconeogenic program is upregulated in L-Δprkar1a as compared to prkar1afl/fl mice; (bottom) gluconeogenic program is downregulated in D-glucose as compared to saline-mice (mean±SEM, * P<0.05)..
E GSIS of WT mouse islets cultured in serum free media conditioned with plasma of prkar1afl/fl or L-Δprkar1a mice. Prkar1afl/fl plasma does not affect GSIS. L-Δprkar1a plasma at 1:10 but not at 1:100 dilution suppresses GSIS (mean±SEM, * P<0.05).
F Volcano plot of gene expression analysis of liver from prkar1afl/fl and L-Δprkar1a mice. Significant upregulation of Kiss1 transcript is detected in L-Δprkar1a mice.
G (top) qRT-PCR of Kiss1 transcript and (bottom) IB in liver tissue from mice with indicated liver genetic complement or intravenous infusion. L-Δprkar1a liver shows increased Kiss1 transcript and kisspeptin protein. D-glucose mice show Kiss1 downregulation as compared to controls (mean±SEM, * P<0.05).