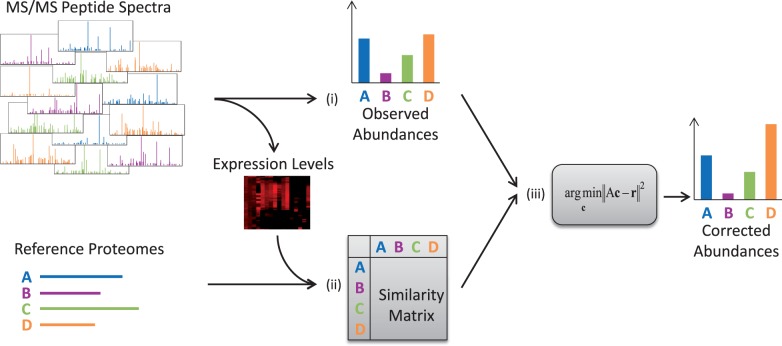
Fig. 1.
Method overview. Pipasic involves three main steps: (i) peptide identification: here metaproteomic peptide spectra are identified by a database search. The number of matches to the proteomes is the observed abundances. (ii) Similarity estimation: the similarities between the reference proteomes are calculated and stored in a similarity matrix. This incorporates the adjustment to only regard expressed proteins and to weight them according to their expression level. (iii) Similarity correction: the observed abundances are corrected using the similarity matrix yielding corrected abundances