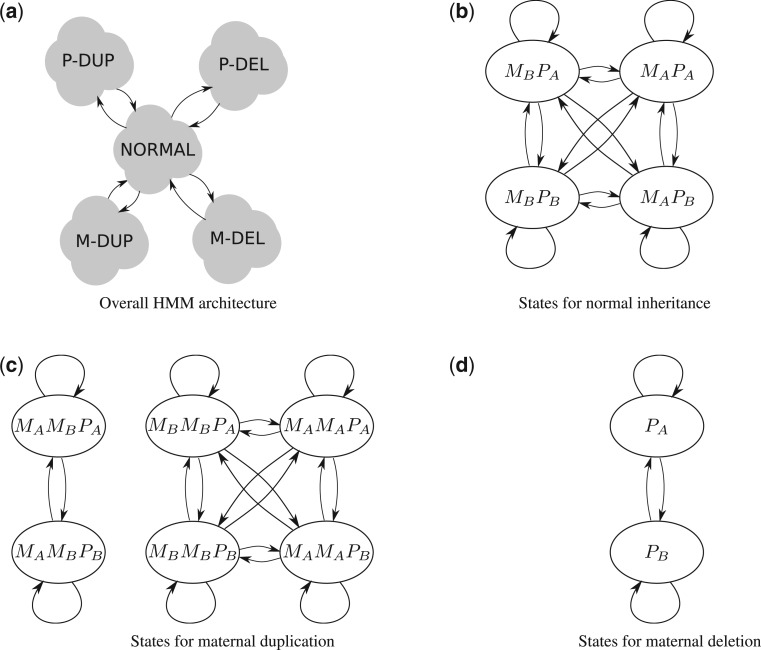
Fig. 3.
HMM used for CNV inference. (a) High-level architecture of the HMM with five sets of states corresponding to five types of fetal inheritance. Note, we do not allow two CNVs to be adjacent; thus, switching between two CNVs always has to go through a normal inheritance state. Edges in (a) represent edges coming in/out of all states between two sets of states. (b–d) Correspond to the diagram of states of the HMM within the normal inheritance, maternal duplication and maternal deletion states of (a). Paternal duplications/deletions are analogous to (c) and (d). Inner edges in (b–d) serve to model errors in phasing or recombination events