Fig. 1.
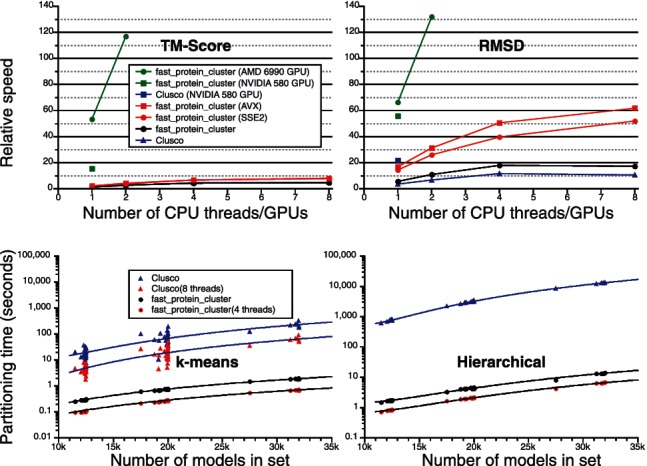
Performance of fast_protein_cluster. The speeds of all-atom RMSD and TM-score matrix calculations over the entire Spicker test set are shown relative to qcprot and the original TM-score for the different methodologies on a 4-core I7 CPU and two different GPUs. The times for k-means and hierarchical partitioning are shown as a function of the number of models. For RMSD calculations, the parallel SSE2 and AVX SIMD code on the laptop CPU outperform the Clusco GPU code. For partitioning, fast_protein_cluster is up to 250× and 2000× faster for k-means and hierarchical clustering, respectively
