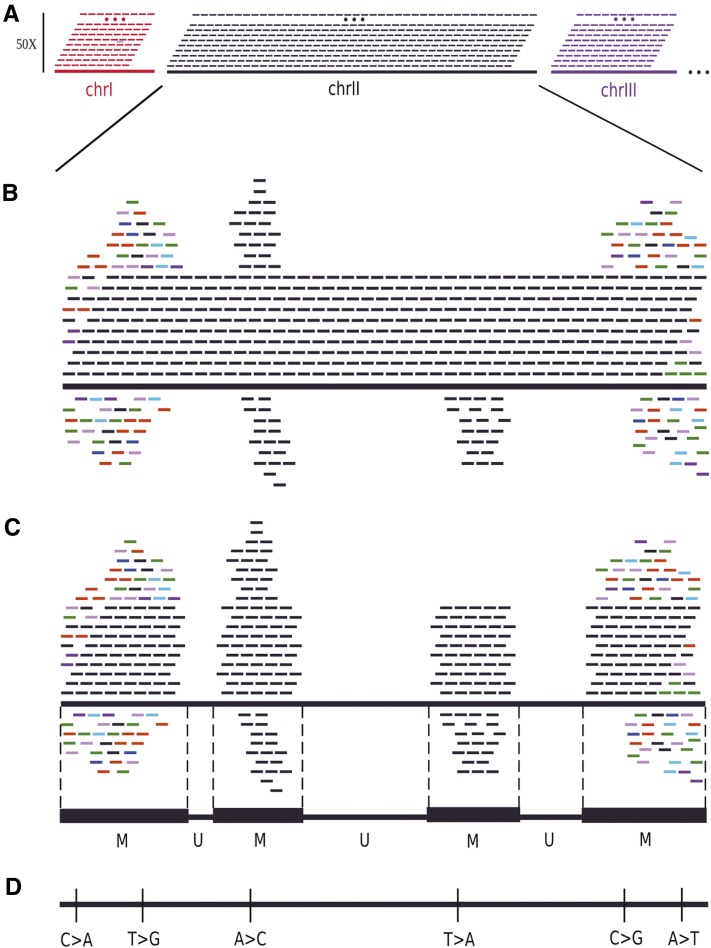
Figure 1.
Principle of virtual HTS profiling of a reference genome. (A) Virtual read production. A read is generated at each genomic position on the forward strand so that the coverage reaches the read length (50× in this example). The next steps are illustrated with the chromosome II. (B) Read mapping. Although reads were only generated on the forward strand, some of them, coming from repetitive regions within the same chromosome (black reads) or different chromosomes (colored reads) exhibit multialignments and/or are aligned on the reverse strand. (C) Annotation of the M and U regions. The start and end coordinates of continuous M regions are reported after unique alignments were discarded. Gaps between M regions are converted to U regions. (D) Intragenomic SNV identification. The mismatches analysis in M regions allowed identification of the indicated SNVs found in repetitive regions of the genome. HTS, high-throughput sequencing; SNV, single-nucleotide variant.