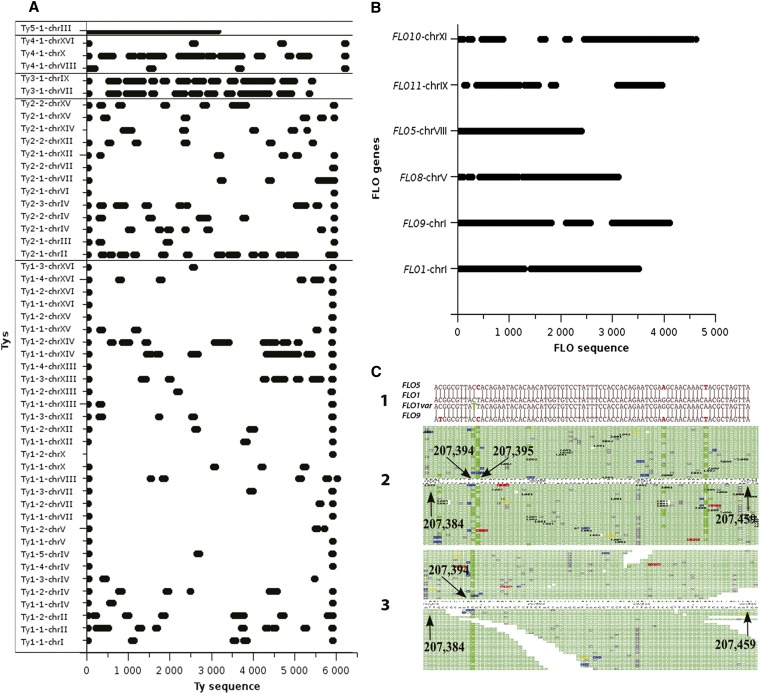
Figure 6.
Analysis of M regions from two classes of repetitive elements. (A) MU regions overlapping the Ty1-Ty5 elements. (B) MU regions overlapping the six FLO gene copies. The MU regions are represented by horizontal bold bars. (C) Detection of acquired mutations within the polymorphic FLO1, FLO5, and FLO9 multi-copy genes. (1) Alignment of the FLO5, FLO1, FLO9, and FLO1 variant (identified here) gene sequences. (2) Visualization of the forward and reverse strand read mapping of the FLO1 gene on the chromosome I in the experimental HTS from the wt_A100 wild type line with the SOLiD Alignment Browser. The coordinates of the regions are indicated (arrows). The fluorescent green bars outline positions of mismatches consistent with a base change. (3) Same as (2) after using g-deNoise. The reads containing the intragenomic SNVs were removed and the unpredicted (acquired) SNP at position 207,394 persisted. The nature and position of the acquired mutation is indicated in green in the FLO1 variant sequence in (1). This identification of this acquired SNP was based on the examination of the 50 nt-reads, containing the adjacent FLO1 specific polymorphisms. It was also confirmed by Sanger sequencing of a polymerase chain reaction product amplified using primers designed in the U regions flanking the FLO1 gene. SNP, single-nucleotide polymorphism.