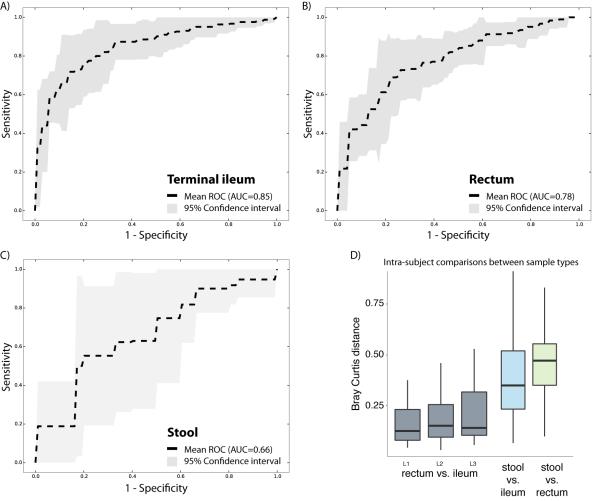
Figure 4. Disease classification performs well on biopsy-associated microbiome profiles.
(A-C) For each of the three sample types, including terminal ileum biopsy (A), rectum biopsy (B), and stool sample (C), we evaluated the accuracy of disease classification using L1 penalized logistic regression with ROC curves representing the results. Dashed lines show the mean performance obtained when genus-level features were used, and the surrounding grey area is the 95% confidence interval. Terminal ileum biopsies performed best (AUC = 0.85), closely matched by the rectum biopsies (AUC = 0.78). However, the classifier based on the stool samples collected at the time of the diagnosis performs less well (AUC = 0.66), and with low consistency (large confidence interval). (D) The intra-subject diversity in microbiome composition was determined for all pairwise sample type combinations. Both biopsy samples were found to be highly similar, whereas the stool sample was quite diverse. Further, we also compared whether disease location would impact the intra-subject diversity between the two tissue biopsy locations. The location of the disease, ileal (L1), colonic (L2), or ileocolonic (L3), did not significantly disrupt the similarity between the two intra-subject mucosal-associated microbiota. Also, no biomarker was detected allowing us to distinguish these disease sub-phenotypes. Related to Figure S3.