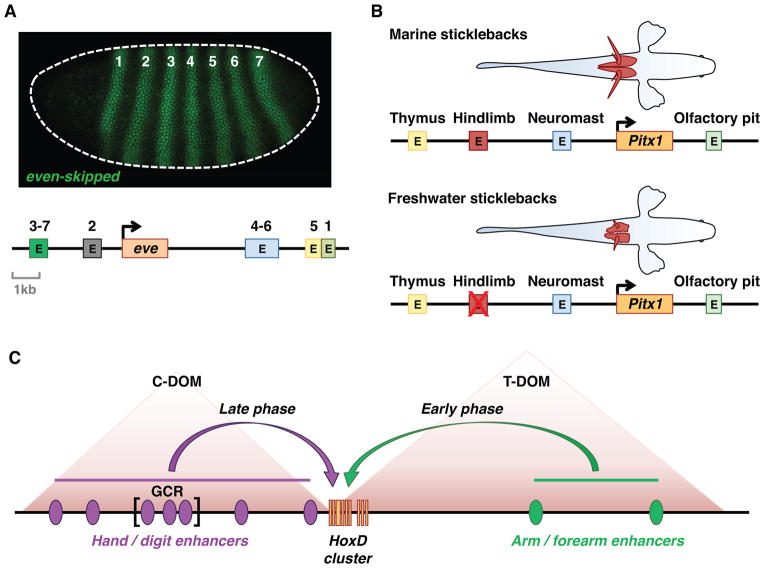
Figure 1. Organization of cis-regulatory DNAs in metazoan genomes.
Metazoan genes are regulated by multiple enhancers. (A) Organization of the even-skipped (eve) locus in the Drosophila genome. The eve gene is just 3 kb in length, but is regulated by individual stripe enhancers (E) located in both 5′ and 3′ flanking regions. The eve stripe enhancers function in an additive fashion to produce 7 stripes of gene expression in the early Drosophila embryo (micrograph by Mike Perry and Michael Levine, personal communication). (B) Evolution of pelvic fins in stickleback fish. The Pitx1 gene is regulated in different tissues by a series of enhancers located in both 5′ and 3′ flanking regions. Deletion of the hindlimb enhancer results in reduced development of the pelvic fins (red) in freshwater populations (adapted from (Shapiro et al., 2004)). (C) Organization of the HoxD complex in mice. The complex is regulated by a series of flanking enhancers (purple and green ovals) located in two neighboring topological association domains (TADs). The telomeric TAD (T-DOM) regulates linked HoxD genes in the developing arm and forearm, while the centromeric TAD (C-DOM) regulates expression in the hand and the digits (adapted from Andrey et al., 2013).